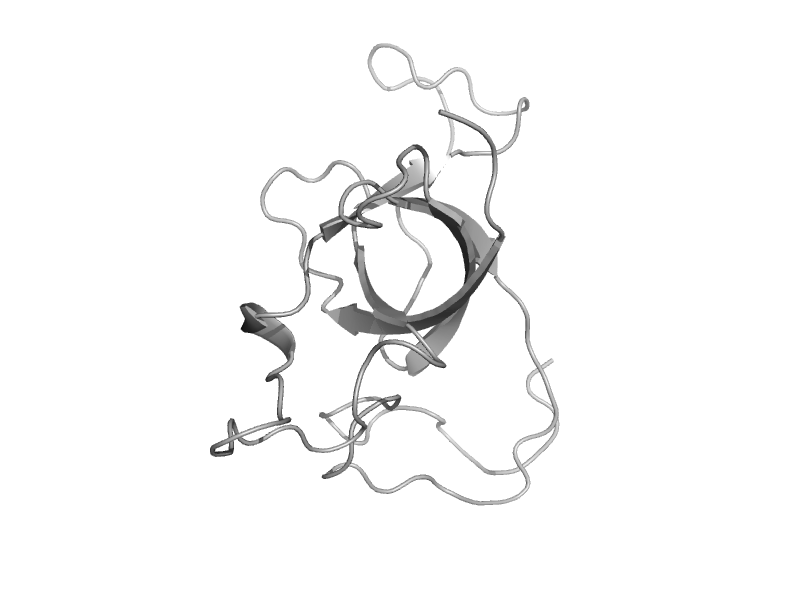
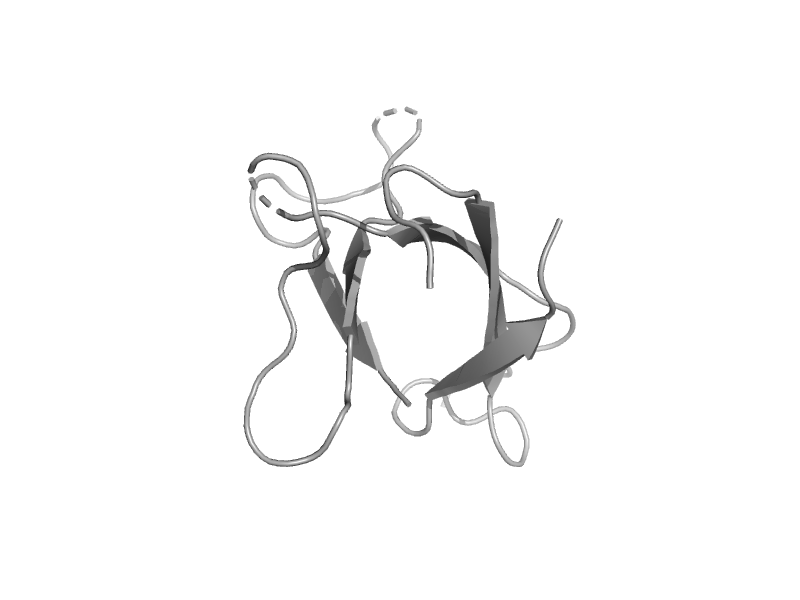
solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1k28a1 ( 6 ) nnlnwFvGVVEdr---mDplklGrVrVRVv-glhppqraqgdvmGipt
d2p5zx1 ( 378 ) rpvmagtlpArVtsdiyahid-kdgryrVnLdfdtwk-----------pg
bbbbb bbbb
60 70 80 90 100
d1k28a1 ( 50 ) ekLpwMsviqpitSaamsgiggsvtgpvegtrVyGhFldkwktnGiVlgt
d2p5zx1 ( 422 ) yeslwVrl-------------------lAgteVsIafeegnpdrpyiagv
bb bbbbbb bbbbbb
110 120 130
d1k28a1 ( 100 ) yggivrekpnrleGfsDptgqyprrlgndt
d2p5zx1 ( 468 ) k
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | N | N | L | N | W | F | V | G | V | V | E | D | R | - | - | - | M | D | P | L | K | L | G | R | V | R | V | R | V | V | - | G | L | H | P | P | Q | R | A | Q | G | D | V | M | G | I | P | T | E | K | L | P | W | M | S | V | I | Q | P | I | T | S | A | A | M | S | G | I | G | G | S | V | T | G | P | V | E | G | T | R | V | Y | G | H | F | L | D | K | W | K | T | N | G | I | V | L | G | T | Y | G | G | I | V | R | E | K | P | N | R | L | E | G | F | S | D | P | T | G | Q | Y | P | R | R | L | G | N | D | T |
| R | P | V | M | A | G | T | L | P | A | R | V | T | S | D | I | Y | A | H | I | D | - | K | D | G | R | Y | R | V | N | L | D | F | D | T | W | K | - | - | - | - | - | - | - | - | - | - | - | P | G | Y | E | S | L | W | V | R | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | A | G | T | E | V | S | I | A | F | E | E | G | N | P | D | R | P | Y | I | A | G | V | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1k28a1 | A | P16009 | GO:0003796 |
| d1k28a1 | A | P16009 | GO:0005515 |
| d1k28a1 | A | P16009 | GO:0009253 |
| d1k28a1 | A | P16009 | GO:0016798 |
| d1k28a1 | A | P16009 | GO:0016998 |
| d1k28a1 | A | P16009 | GO:0031640 |
| d1k28a1 | A | P16009 | GO:0042742 |
| d1k28a1 | A | P16009 | GO:0042802 |
| d1k28a1 | A | P16009 | GO:0044409 |
| d1k28a1 | A | P16009 | GO:0046718 |
| d1k28a1 | A | P16009 | GO:0098003 |
| d1k28a1 | A | P16009 | GO:0098015 |
| d1k28a1 | A | P16009 | GO:0098025 |
| d1k28a1 | A | P16009 | GO:0098932 |
| d1k28a1 | A | P16009 | GO:0098994 |
| d1k28a1 | A | P17172 | GO:0003796 |
| d1k28a1 | A | P17172 | GO:0005515 |
| d1k28a1 | A | P17172 | GO:0009253 |
| d1k28a1 | A | P17172 | GO:0016798 |
| d1k28a1 | A | P17172 | GO:0016998 |
| d1k28a1 | A | P17172 | GO:0031640 |
| d1k28a1 | A | P17172 | GO:0042742 |
| d1k28a1 | A | P17172 | GO:0042802 |
| d1k28a1 | A | P17172 | GO:0044409 |
| d1k28a1 | A | P17172 | GO:0046718 |
| d1k28a1 | A | P17172 | GO:0098003 |
| d1k28a1 | A | P17172 | GO:0098015 |
| d1k28a1 | A | P17172 | GO:0098025 |
| d1k28a1 | A | P17172 | GO:0098932 |
| d1k28a1 | A | P17172 | GO:0098994 |