solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1ek9a- ( 1 ) enLqVYqqA
d1yc9a- ( 63 ) wPsanWWqrYqdaqLnhLIeeA
d3pika- ( 1 ) cslapdyqrpampVpqqFs--------lyqNagwrtFFvdnqVktlIseA
d5azpa- ( 1 ) ctvgpdyrtpdtaaAkidaTaskpYdrsrfeslWWkqFddptLnqLVeqS
d5azsa- ( 1 ) csmaptyerpaapvadsWs------------ldwksFIvdaeLrrLVdmA
aaaaaaaaaa
60 70 80 90 100
d1ek9a- ( 11 ) rlsNpelrksaadrdaafekinearspllpqlglgadytysngyr-----
d1yc9a- ( 85 ) lqhSpslcmamarlkgaqgfarqagairsfdlglaasateskvseryqsa
d3pika- ( 54 ) lvnNrdlrmatlkvqearaqyrltdadrypqlngegsgswsgnl------
d5azpa- ( 51 ) lsgNrdlrvafarLraaralrddvAndrfpvvtsrasadigkgqqpg--v
d5azsa- ( 52 ) LdnnrsLrqtlldieaaraqyriqradrvpglnaaatgnrqrqpad-lsa
aaa aaaaaaaaaaaaaaaaaaaaa bbbbbbbbbbbb
110 120 130 140 150
d1ek9a- ( 56 ) ----danginsnatsaslqlt-qSi--fdskwraltlqekaAgiqdVtyq
d1yc9a- ( 135 ) tppd----gwndyGtltlnfq-ydfdfwgkNraavvaAtSelaAaeaesv
d3pika- ( 98 ) kgnt----attrefstglnas-fdldffGrlkNmSeaerQnylateeaqr
d5azpa- ( 99 ) tedr----vnserydlgldsa-weldlfGrIrRqlessdAlseAaeAdlq
d5azsa- ( 101 ) gnrs----evassyqvglalpeyeldlfGrVkSlTdaalqqylAseeaar
bbbbbbbbbbbb bb aaaaaaaaaaaaaaaaaaaaa
160 170 180 190 200
d1ek9a- ( 100 ) tdqqtLIlnTAtaYFnVLnaidvLsyTqaqkeaIyrqLdqTtqrfnvglv
d1yc9a- ( 180 ) aarlmISTsIAnaYAeLarlyaNqetVhaalqvRnktvelLekryangle
d3pika- ( 143 ) avhilLvSnVAqsYFnQQlAyaQlqiAeetlrnYqqsyafVekqlltgss
d5azpa- ( 144 ) qlqVsLIAeLVdaYGqLRGAqlrekiAlsNlenQkesrqlTeqlrdagvg
d5azsa- ( 147 ) aariaLVAeVSqaYLsYDGAlrrlalTrqtlvsReysfalidqrraagaA
aaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaa
210 220 230 240 250
d1ek9a- ( 150 ) aitdvqnAraqydtVlaneltarnnLdnaveqLrqiTg-------nyype
d1yc9a- ( 230 ) tlgsVsqAkavaasveaellgiqesIqlqknaLAalVgqgpdrAasIeeP
d3pika- ( 193 ) nvlAleqArgviesTrsdiakrqgelaqAnnaLqllLgs-y---gklpqa
d5azpa- ( 194 ) aeldVlrAdarlaaTaasvpqLqaeAerarhrIAtLLgqrpeeltvdLsp
d5azsa- ( 197 ) taldyqealgLVeqAraeqerNlrqkqqafnaLvllLgs-ddAaqAIprs
aaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaa
260 270 280 290 300
d1ek9a- ( 193 ) LAaLnvenfktdkpqpvnallkeAekrnlsLlqarlsqdlAreqirqaqD
d1yc9a- ( 280 ) h--it----ltsryv-------gllghRaditAarwraeAAaqqvgiaqa
d3pika- ( 239 ) qtvns--dsLqsvklpaglsSqiLl-qrpdImeAehaLmaanAnIGaarA
d5azpa- ( 244 ) rdlpa----ItkaLP--IgdPgeLlrrRPdIraaerrlaAstAdVGvatA
d5azsa- ( 246 ) pgqr---pKLlqdiap--gtPseLierrPdIlaaehrLrArnadiGaarA
aaaaaaaaaaaaaaaaaaaaaa
310 320 330 340 350
d1ek9a- ( 243 ) ghlptldltastgisdtsysgsktrgaag---tqyddsngqnkvglsfsl
d1yc9a- ( 324 ) qfypdvtlsafigyqaf------------gldhlfdsgndagaigpaiyl
d3pika- ( 286 ) affpsisltsgistass------------dlsslfnassgmwnfipkiei
d5azpa- ( 288 ) dlfprvslsgflgftag------------rgsqigssaArawsvgpsisw
d5azsa- ( 291 ) affprisltgsfgtssa------------emsglfdggsrswsflptltl
bbbbbbbbbbbb bbbbbbbbbb
360 370 380 390 400
d1ek9a- ( 291 ) piyqgg-vnsqvkqaqynfvgaseqlesahrsVvqtVrsSfnnInasiss
d1yc9a- ( 362 ) plftggrlegqltsaearyqeAvaqyngtlvqAlheiadvvtssqalqar
d3pika- ( 324 ) pifnagrnqanldiaeirqqqsvvnYeqkIqnAfkeVadAlalrqslndq
d5azpa- ( 326 ) aafdlgsVrarlrgakadadaalasYeqqvllAleeSanaFsdYgkrqer
d5azsa- ( 329 ) pifdggrnranlslaearkdsavaaYegtIqtAfreVadalaasdtlrre
aaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaa
410 420 430 440 450
d1ek9a- ( 341 ) Inaykqavvsaqssld-aeagysvgtrtIvdVLdaTttLynakqelAnar
d1yc9a- ( 412 ) inkteqavqqaeqalhiatnrYqgglatYldVlvaEesllnnqralvnlq
d3pika- ( 374 ) IsaqqrylaslqitlqraralyqhgavsYleVldaErslfaTrqtlldln
d5azpa- ( 376 ) lvslvrqSeaSraaaqqaairYregttdFlvLldaereqlsAedaqAqAe
d5azsa- ( 379 ) ekalralAnssnealklakaryesgvdnHlryLdaqrssflneiafidgs
aaaaaaaaaaaaaaaaaaaaaaa aaaaaaaaaaaaaaaaaaaaaa
460 470 480
d1ek9a- ( 391 ) YnYlinqLniksalGtLneqdllalnnaLskpvsTnpe
d1yc9a- ( 462 ) sraFsldLalIhAlgGGf
d3pika- ( 424 ) yarqvneIsLYtaLgGG
d5azpa- ( 426 ) velYrGIVaIYrSLGGGwqps
d5azsa- ( 429 ) tqrqiaLVdLfrAlG
aaaaaaaaaaaaaa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Integral outer membrane protein TolC, efflux pump component | Outer membrane efflux proteins (OEP) | Escherichia coli [TaxId: 562] | 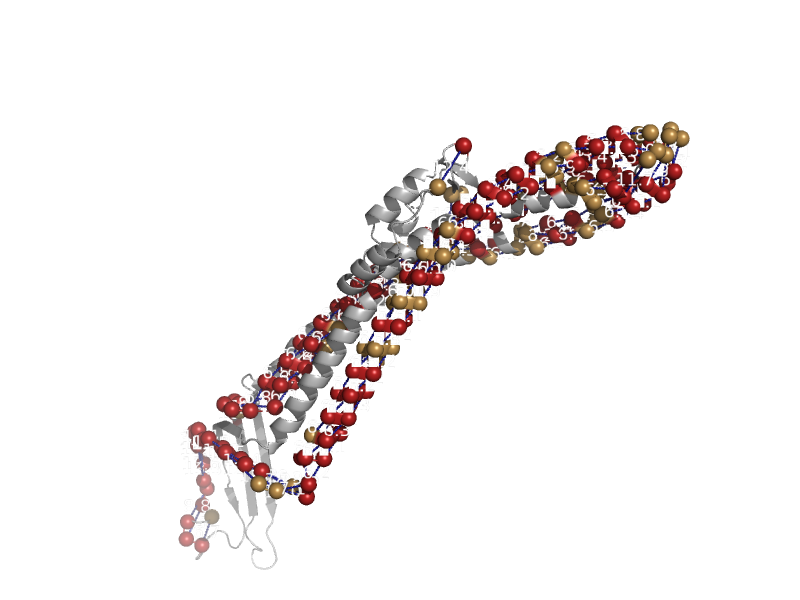 | view | |
| automated matches | automated matches | Vibrio cholerae [TaxId: 666] |  | view | |
| automated matches | automated matches | Escherichia coli [TaxId: 562] | 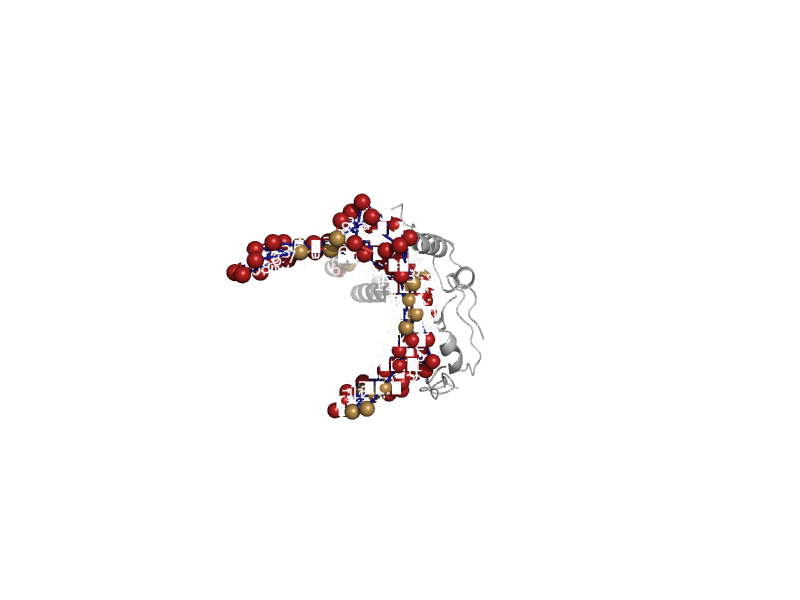 | view | |
| automated matches | automated matches | Pseudomonas aeruginosa [TaxId: 208964] | 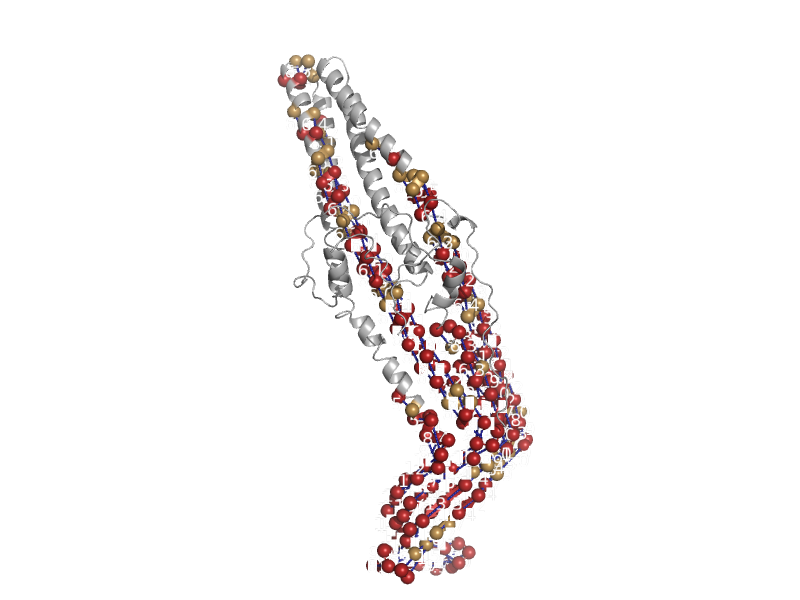 | view | |
| automated matches | automated matches | Pseudomonas aeruginosa [TaxId: 208964] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | 330 | 340 | 350 | 360 | 370 | 380 | 390 | 400 | 410 | 420 | 430 | 440 | 450 | 460 | 470 | 480 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | N | L | Q | V | Y | Q | Q | A | R | L | S | N | P | E | L | R | K | S | A | A | D | R | D | A | A | F | E | K | I | N | E | A | R | S | P | L | L | P | Q | L | G | L | G | A | D | Y | T | Y | S | N | G | Y | R | - | - | - | - | - | - | - | - | - | D | A | N | G | I | N | S | N | A | T | S | A | S | L | Q | L | T | - | Q | S | I | - | - | F | D | S | K | W | R | A | L | T | L | Q | E | K | A | A | G | I | Q | D | V | T | Y | Q | T | D | Q | Q | T | L | I | L | N | T | A | T | A | Y | F | N | V | L | N | A | I | D | V | L | S | Y | T | Q | A | Q | K | E | A | I | Y | R | Q | L | D | Q | T | T | Q | R | F | N | V | G | L | V | A | I | T | D | V | Q | N | A | R | A | Q | Y | D | T | V | L | A | N | E | L | T | A | R | N | N | L | D | N | A | V | E | Q | L | R | Q | I | T | G | - | - | - | - | - | - | - | N | Y | Y | P | E | L | A | A | L | N | V | E | N | F | K | T | D | K | P | Q | P | V | N | A | L | L | K | E | A | E | K | R | N | L | S | L | L | Q | A | R | L | S | Q | D | L | A | R | E | Q | I | R | Q | A | Q | D | G | H | L | P | T | L | D | L | T | A | S | T | G | I | S | D | T | S | Y | S | G | S | K | T | R | G | A | A | G | - | - | - | T | Q | Y | D | D | S | N | G | Q | N | K | V | G | L | S | F | S | L | P | I | Y | Q | G | G | - | V | N | S | Q | V | K | Q | A | Q | Y | N | F | V | G | A | S | E | Q | L | E | S | A | H | R | S | V | V | Q | T | V | R | S | S | F | N | N | I | N | A | S | I | S | S | I | N | A | Y | K | Q | A | V | V | S | A | Q | S | S | L | D | - | A | E | A | G | Y | S | V | G | T | R | T | I | V | D | V | L | D | A | T | T | T | L | Y | N | A | K | Q | E | L | A | N | A | R | Y | N | Y | L | I | N | Q | L | N | I | K | S | A | L | G | T | L | N | E | Q | D | L | L | A | L | N | N | A | L | S | K | P | V | S | T | N | P | E |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | W | P | S | A | N | W | W | Q | R | Y | Q | D | A | Q | L | N | H | L | I | E | E | A | L | Q | H | S | P | S | L | C | M | A | M | A | R | L | K | G | A | Q | G | F | A | R | Q | A | G | A | I | R | S | F | D | L | G | L | A | A | S | A | T | E | S | K | V | S | E | R | Y | Q | S | A | T | P | P | D | - | - | - | - | G | W | N | D | Y | G | T | L | T | L | N | F | Q | - | Y | D | F | D | F | W | G | K | N | R | A | A | V | V | A | A | T | S | E | L | A | A | A | E | A | E | S | V | A | A | R | L | M | I | S | T | S | I | A | N | A | Y | A | E | L | A | R | L | Y | A | N | Q | E | T | V | H | A | A | L | Q | V | R | N | K | T | V | E | L | L | E | K | R | Y | A | N | G | L | E | T | L | G | S | V | S | Q | A | K | A | V | A | A | S | V | E | A | E | L | L | G | I | Q | E | S | I | Q | L | Q | K | N | A | L | A | A | L | V | G | Q | G | P | D | R | A | A | S | I | E | E | P | H | - | - | I | T | - | - | - | - | L | T | S | R | Y | V | - | - | - | - | - | - | - | G | L | L | G | H | R | A | D | I | T | A | A | R | W | R | A | E | A | A | A | Q | Q | V | G | I | A | Q | A | Q | F | Y | P | D | V | T | L | S | A | F | I | G | Y | Q | A | F | - | - | - | - | - | - | - | - | - | - | - | - | G | L | D | H | L | F | D | S | G | N | D | A | G | A | I | G | P | A | I | Y | L | P | L | F | T | G | G | R | L | E | G | Q | L | T | S | A | E | A | R | Y | Q | E | A | V | A | Q | Y | N | G | T | L | V | Q | A | L | H | E | I | A | D | V | V | T | S | S | Q | A | L | Q | A | R | I | N | K | T | E | Q | A | V | Q | Q | A | E | Q | A | L | H | I | A | T | N | R | Y | Q | G | G | L | A | T | Y | L | D | V | L | V | A | E | E | S | L | L | N | N | Q | R | A | L | V | N | L | Q | S | R | A | F | S | L | D | L | A | L | I | H | A | L | G | G | G | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| C | S | L | A | P | D | Y | Q | R | P | A | M | P | V | P | Q | Q | F | S | - | - | - | - | - | - | - | - | L | Y | Q | N | A | G | W | R | T | F | F | V | D | N | Q | V | K | T | L | I | S | E | A | L | V | N | N | R | D | L | R | M | A | T | L | K | V | Q | E | A | R | A | Q | Y | R | L | T | D | A | D | R | Y | P | Q | L | N | G | E | G | S | G | S | W | S | G | N | L | - | - | - | - | - | - | K | G | N | T | - | - | - | - | A | T | T | R | E | F | S | T | G | L | N | A | S | - | F | D | L | D | F | F | G | R | L | K | N | M | S | E | A | E | R | Q | N | Y | L | A | T | E | E | A | Q | R | A | V | H | I | L | L | V | S | N | V | A | Q | S | Y | F | N | Q | Q | L | A | Y | A | Q | L | Q | I | A | E | E | T | L | R | N | Y | Q | Q | S | Y | A | F | V | E | K | Q | L | L | T | G | S | S | N | V | L | A | L | E | Q | A | R | G | V | I | E | S | T | R | S | D | I | A | K | R | Q | G | E | L | A | Q | A | N | N | A | L | Q | L | L | L | G | S | - | Y | - | - | - | G | K | L | P | Q | A | Q | T | V | N | S | - | - | D | S | L | Q | S | V | K | L | P | A | G | L | S | S | Q | I | L | L | - | Q | R | P | D | I | M | E | A | E | H | A | L | M | A | A | N | A | N | I | G | A | A | R | A | A | F | F | P | S | I | S | L | T | S | G | I | S | T | A | S | S | - | - | - | - | - | - | - | - | - | - | - | - | D | L | S | S | L | F | N | A | S | S | G | M | W | N | F | I | P | K | I | E | I | P | I | F | N | A | G | R | N | Q | A | N | L | D | I | A | E | I | R | Q | Q | Q | S | V | V | N | Y | E | Q | K | I | Q | N | A | F | K | E | V | A | D | A | L | A | L | R | Q | S | L | N | D | Q | I | S | A | Q | Q | R | Y | L | A | S | L | Q | I | T | L | Q | R | A | R | A | L | Y | Q | H | G | A | V | S | Y | L | E | V | L | D | A | E | R | S | L | F | A | T | R | Q | T | L | L | D | L | N | Y | A | R | Q | V | N | E | I | S | L | Y | T | A | L | G | G | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| C | T | V | G | P | D | Y | R | T | P | D | T | A | A | A | K | I | D | A | T | A | S | K | P | Y | D | R | S | R | F | E | S | L | W | W | K | Q | F | D | D | P | T | L | N | Q | L | V | E | Q | S | L | S | G | N | R | D | L | R | V | A | F | A | R | L | R | A | A | R | A | L | R | D | D | V | A | N | D | R | F | P | V | V | T | S | R | A | S | A | D | I | G | K | G | Q | Q | P | G | - | - | V | T | E | D | R | - | - | - | - | V | N | S | E | R | Y | D | L | G | L | D | S | A | - | W | E | L | D | L | F | G | R | I | R | R | Q | L | E | S | S | D | A | L | S | E | A | A | E | A | D | L | Q | Q | L | Q | V | S | L | I | A | E | L | V | D | A | Y | G | Q | L | R | G | A | Q | L | R | E | K | I | A | L | S | N | L | E | N | Q | K | E | S | R | Q | L | T | E | Q | L | R | D | A | G | V | G | A | E | L | D | V | L | R | A | D | A | R | L | A | A | T | A | A | S | V | P | Q | L | Q | A | E | A | E | R | A | R | H | R | I | A | T | L | L | G | Q | R | P | E | E | L | T | V | D | L | S | P | R | D | L | P | A | - | - | - | - | I | T | K | A | L | P | - | - | I | G | D | P | G | E | L | L | R | R | R | P | D | I | R | A | A | E | R | R | L | A | A | S | T | A | D | V | G | V | A | T | A | D | L | F | P | R | V | S | L | S | G | F | L | G | F | T | A | G | - | - | - | - | - | - | - | - | - | - | - | - | R | G | S | Q | I | G | S | S | A | A | R | A | W | S | V | G | P | S | I | S | W | A | A | F | D | L | G | S | V | R | A | R | L | R | G | A | K | A | D | A | D | A | A | L | A | S | Y | E | Q | Q | V | L | L | A | L | E | E | S | A | N | A | F | S | D | Y | G | K | R | Q | E | R | L | V | S | L | V | R | Q | S | E | A | S | R | A | A | A | Q | Q | A | A | I | R | Y | R | E | G | T | T | D | F | L | V | L | L | D | A | E | R | E | Q | L | S | A | E | D | A | Q | A | Q | A | E | V | E | L | Y | R | G | I | V | A | I | Y | R | S | L | G | G | G | W | Q | P | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| C | S | M | A | P | T | Y | E | R | P | A | A | P | V | A | D | S | W | S | - | - | - | - | - | - | - | - | - | - | - | - | L | D | W | K | S | F | I | V | D | A | E | L | R | R | L | V | D | M | A | L | D | N | N | R | S | L | R | Q | T | L | L | D | I | E | A | A | R | A | Q | Y | R | I | Q | R | A | D | R | V | P | G | L | N | A | A | A | T | G | N | R | Q | R | Q | P | A | D | - | L | S | A | G | N | R | S | - | - | - | - | E | V | A | S | S | Y | Q | V | G | L | A | L | P | E | Y | E | L | D | L | F | G | R | V | K | S | L | T | D | A | A | L | Q | Q | Y | L | A | S | E | E | A | A | R | A | A | R | I | A | L | V | A | E | V | S | Q | A | Y | L | S | Y | D | G | A | L | R | R | L | A | L | T | R | Q | T | L | V | S | R | E | Y | S | F | A | L | I | D | Q | R | R | A | A | G | A | A | T | A | L | D | Y | Q | E | A | L | G | L | V | E | Q | A | R | A | E | Q | E | R | N | L | R | Q | K | Q | Q | A | F | N | A | L | V | L | L | L | G | S | - | D | D | A | A | Q | A | I | P | R | S | P | G | Q | R | - | - | - | P | K | L | L | Q | D | I | A | P | - | - | G | T | P | S | E | L | I | E | R | R | P | D | I | L | A | A | E | H | R | L | R | A | R | N | A | D | I | G | A | A | R | A | A | F | F | P | R | I | S | L | T | G | S | F | G | T | S | S | A | - | - | - | - | - | - | - | - | - | - | - | - | E | M | S | G | L | F | D | G | G | S | R | S | W | S | F | L | P | T | L | T | L | P | I | F | D | G | G | R | N | R | A | N | L | S | L | A | E | A | R | K | D | S | A | V | A | A | Y | E | G | T | I | Q | T | A | F | R | E | V | A | D | A | L | A | A | S | D | T | L | R | R | E | E | K | A | L | R | A | L | A | N | S | S | N | E | A | L | K | L | A | K | A | R | Y | E | S | G | V | D | N | H | L | R | Y | L | D | A | Q | R | S | S | F | L | N | E | I | A | F | I | D | G | S | T | Q | R | Q | I | A | L | V | D | L | F | R | A | L | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1ek9a_ | A | P02930 | GO:0005216 |
| d1ek9a_ | A | P02930 | GO:0005515 |
| d1ek9a_ | A | P02930 | GO:0009279 |
| d1ek9a_ | A | P02930 | GO:0009410 |
| d1ek9a_ | A | P02930 | GO:0009636 |
| d1ek9a_ | A | P02930 | GO:0014070 |
| d1ek9a_ | A | P02930 | GO:0015125 |
| d1ek9a_ | A | P02930 | GO:0015288 |
| d1ek9a_ | A | P02930 | GO:0015562 |
| d1ek9a_ | A | P02930 | GO:0015721 |
| d1ek9a_ | A | P02930 | GO:0016020 |
| d1ek9a_ | A | P02930 | GO:0019867 |
| d1ek9a_ | A | P02930 | GO:0030288 |
| d1ek9a_ | A | P02930 | GO:0034220 |
| d1ek9a_ | A | P02930 | GO:0042802 |
| d1ek9a_ | A | P02930 | GO:0042930 |
| d1ek9a_ | A | P02930 | GO:0042931 |
| d1ek9a_ | A | P02930 | GO:0046677 |
| d1ek9a_ | A | P02930 | GO:0055085 |
| d1ek9a_ | A | P02930 | GO:0098567 |
| d1ek9a_ | A | P02930 | GO:0140330 |
| d1ek9a_ | A | P02930 | GO:1990196 |
| d1ek9a_ | A | P02930 | GO:1990281 |
| d1ek9a_ | A | P02930 | GO:1990961 |
