solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1ddba- ( 1 ) mdsevsngsglGakhitdllvfgFlqssGctrqelevlGrelpvqAywea
d1k3ka- ( 1 ) mdedvlpgevlaiegiFMacGl----------------------
d1ohua- ( 72 ) dwe-eprLdIegFVVDYFthrirqngewf---------------
d1q59a1 ( 1 ) m-----aystreillAlcirdSrvHGn-g---------------
d2vofa1 ( 2 ) aeselhIhsLAehYLqyVlqvp----afe------------
d3bl2a- ( 5 ) ksgtyWatLItaFLktVSkve-----eldcvd--------
d4bd8a- ( 11 ) ggptsseqimktgalllqgFiqdRagrda------------------
d5vx1a1 ( 23 ) seeqVaqdTeeVFrSYVfyrHqqeqeaegvaapADpeMvt---
d5wdda- ( 21 ) s--ptdkeLVsqAkaLCrdYInsrlira----gvsws----------
d6h1na1 ( 5 ) mscwlreqtlllAedYisfCsgiq----q--------------
d6udva- ( 172 ) dlyrqSleIIsrYLreqAtga----kdtkpMgr-------
d6v4ma- ( 6 ) w--dnelsgintsvsLAaDYVYmrLate----gfvfgirs-------
aaaaaaaaaaaaa
60 70 80 90 100
d1ddba- ( 51 ) dledelqtdgsqasrsfnqgriepdsesqeeiihniArhlAqiGdemdhn
d1k3ka- ( 23 ) -------------nepe----------ylyhpllsPiklyitglmrdke-
d1ohua- ( 101 ) -------------gapg-lps----------gvqpeHervgtifekkha-
d1q59a1 ( 24 ) -------------tlhpvlelaaretplrlspedtvvlrYhvlleeiie-
d2vofa1 ( 30 ) --------------------------------psqAcrVLqrvAfsvQe-
d3bl2a- ( 32 ) ------------------------------saVlvdVskIitlTqefrr-
d4bd8a- ( 55 ) --------------------------------stklseslkriGdelds-
d5vx1a1 ( 63 ) ---------------------------lplqpsstmGqVGrqLAiigdd-
d5wdda- ( 62 ) ------------------------------gklaeVsaiLlrlGdeley-
d6h1na1 ( 30 ) ------------------------------tppsesAeAMrylAkemEq-
d6udva- ( 202 ) ----------------------------sgatSrkAleTLrrvGdgvQr-
d6v4ma- ( 41 ) ----------------------------svrApirLCdaMflMCdlFer-
aaaaaaaaaaaaa
110 120 130 140 150
d1ddba- ( 101 ) iqptlvrqlAaqFmngslseedkrnClAkAldevktaf---prdMenDka
d1k3ka- ( 49 ) ---slFeamlan------vrFhsttgidqlglsmlqvsgdgnmnwgrAla
d1ohua- ( 129 ) ---enFetfSeqLla----vpIsfslYqdvVrtVgn------psygrLiG
d1q59a1 ( 60 ) ---rnsetFtetwnrfithtehvdldfnsvFleifhr---gdpslGrAlA
d2vofa1 ( 48 ) --venlksylddf---hVsidtAriiFnqv-ekefedg---iinWgrIVt
d3bl2a- ( 51 ) hydsvyradygp------aLknWkrdLsklFtslFvd----vinsgrIvg
d4bd8a- ( 73 ) -nmeLqrmia--------avdtsprvffrVaadmfsdg---nfnwgrvVa
d5vx1a1 ( 85 ) inrrydseFqtmLqhaqPtaenAyeyFtkiAtslFes----ginwgrVvA
d5wdda- ( 81 ) irpnvyrnIarqlnIslhsetvVtdaFlavAaqiFta----gitwgkVVs
d6h1na1 ( 49 ) qhrtkfrslSqeFld--tçgadpskçlqsvMrelvgdg---kMnWgrvvs
d6udva- ( 223 ) nhetafqgmlrkldI--kned-dVkslsrvmihvFsdg---vtnWgrIVt
d6v4ma- ( 63 ) fhdryIapLknaclgisad--mdmrmFfsALdsvFss----giswsrIVA
aaaaaaa aaaaaaaaaaa aaaaaa
160 170 180 190 200
d1ddba- ( 148 ) MliMtMllAkkvAs-hap-sllrdvFhttvnFinqnlfSYvrnlvrnemd
d1k3ka- ( 90 ) iltFGSfvAqklsn----ephlrdFAlavlPayAyeAigpqwfrarGgwr
d1ohua- ( 174 ) LiSFGGfVAAkesv---elqgqvrnLfvyTSlFIktri--nWrsWddftl
d1q59a1 ( 104 ) WmAWcMhaCrtlccnqstpyyvvdlsvrgMleaSegld--gwihqqggWs
d2vofa1 ( 92 ) IFAfGGVLLkkLe-------say-kVSsFVAefI-n-nTgewi-qnggwe
d3bl2a- ( 91 ) FFDvGrYVCeevlcpgsw-tedHelLndcMthfFiennLmnhFple
d4bd8a- ( 113 ) lfYFaSklvlkalstk---vpelirtimgwtldflrerllgwiqdqggwd
d5vx1a1 ( 131 ) LLgFGYRLALhVyq-hgl-tgfLgqVTrFVvdFMlhhsIArwIaqrggWv
d5wdda- ( 127 ) LYAVAAgLAvdCvr-haq-pamvhtIVdcLGefVrk-tLvtwLkrrgGWa
d6h1na1 ( 94 ) iFtFtGvlAsells-rg--eegs-rlAetiAdYlGg-ekqdwlvenggWe
d6udva- ( 267 ) lISfgAfVAkhLkt-inq-escIepLAesITdVLVr-tkrdwLvkqrgWd
d6v4ma- ( 108 ) MYAFAGsVAlaCAr-qgr-rqtViaIpewImlYMrr-aIapwIhanggwd
aaaaaaaaaaa aaaaaaaaaaaaa aaaa
210
d1ddba-
d1k3ka- ( 136 ) -glkayctqvlt
d1ohua- ( 228 ) -gqkedyeaea
d1q59a1 ( 152 ) -tliednip-g
d2vofa1 ( 142 ) dgFikkfe
d3bl2a-
d4bd8a- ( 160 ) -glls--yfgtptw
d5vx1a1 ( 179 ) -aAln--------l
d5wdda- ( 174 ) -dItkcvv
d6h1na1 ( 142 ) -GFcrffhna
d6udva- ( 314 ) -gFveffhvedleg
d6v4ma- ( 155 ) -sFIfSqdvlngnh
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Proapoptotic molecule Bid | Bcl-2 inhibitors of programmed cell death | Mouse (Mus musculus) [TaxId: 10090] | 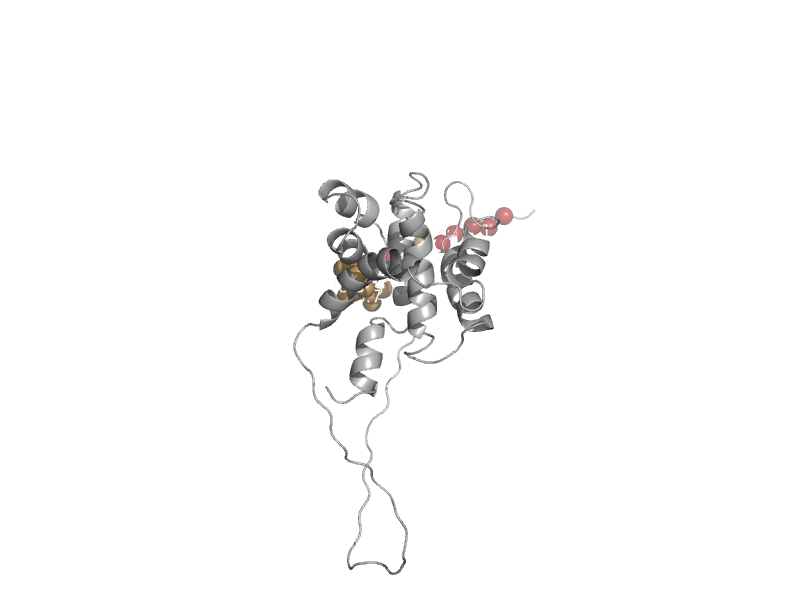 | view | |
| Bcl-2 homolog | Bcl-2 inhibitors of programmed cell death | Kaposi's sarcoma-associated herpesvirus [TaxId: 37296] | 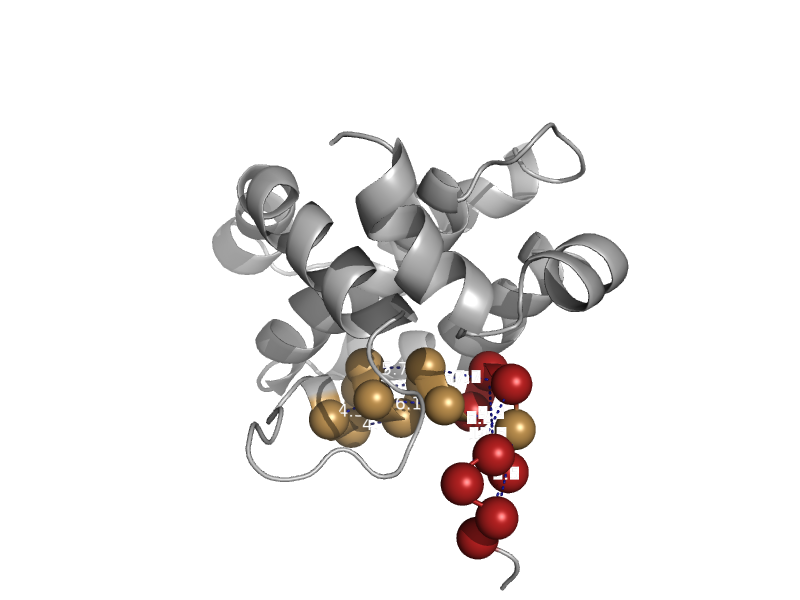 | view | |
| Apoptosis regulator ced-9 | Bcl-2 inhibitors of programmed cell death | Nematode (Caenorhabditis elegans) [TaxId: 6239] | 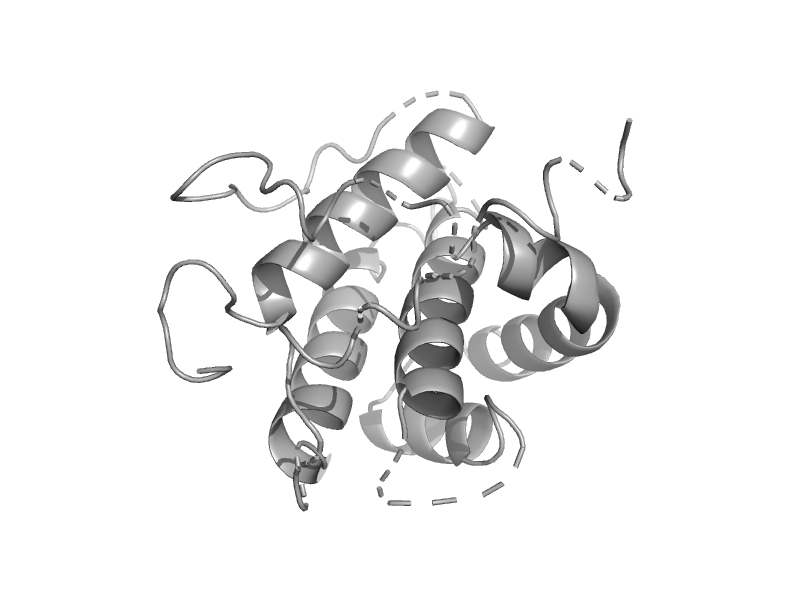 | view | |
| Early antigen protein R | Bcl-2 inhibitors of programmed cell death | Epstein-Barr virus [TaxId: 10376] | 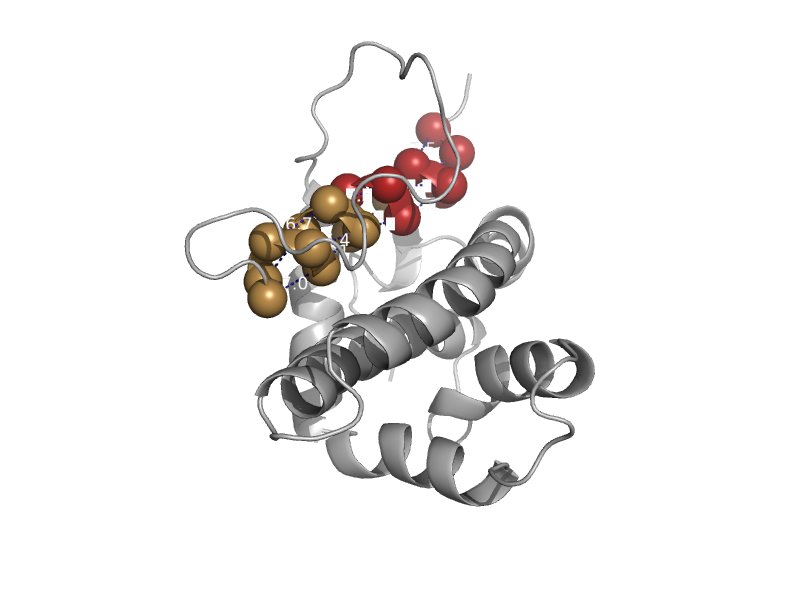 | view | |
| automated matches | automated matches | Mouse (Mus musculus) [TaxId: 10090] | 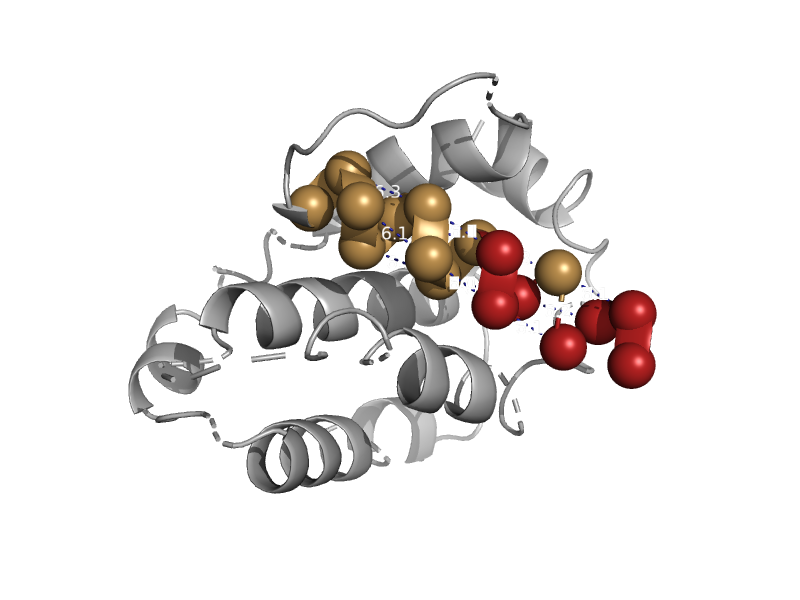 | view | |
| Bcl-2 homolog V-bcl-2 | Bcl-2 inhibitors of programmed cell death | Murid herpesvirus 4 [TaxId: 33708] | 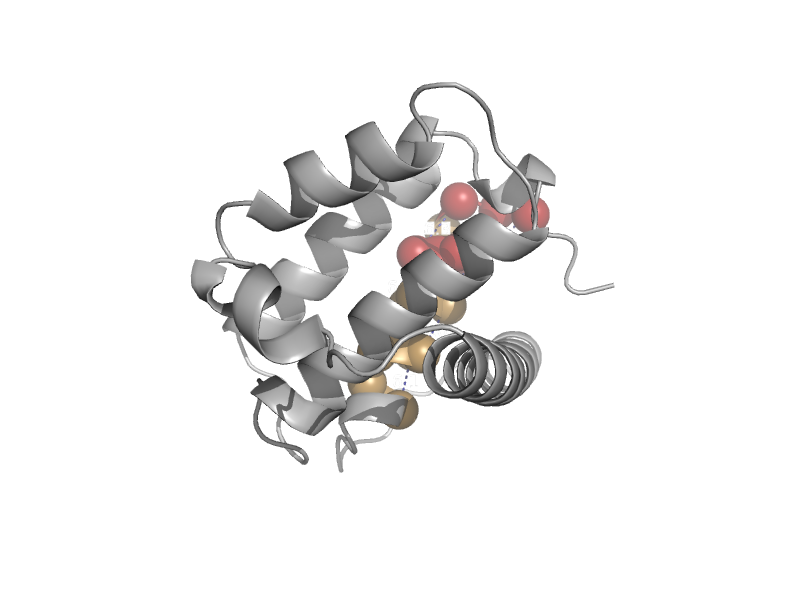 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 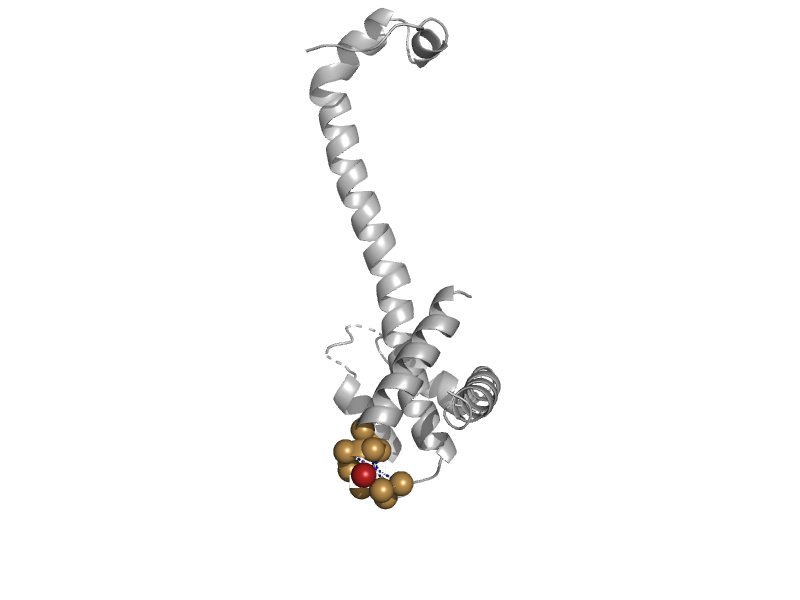 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 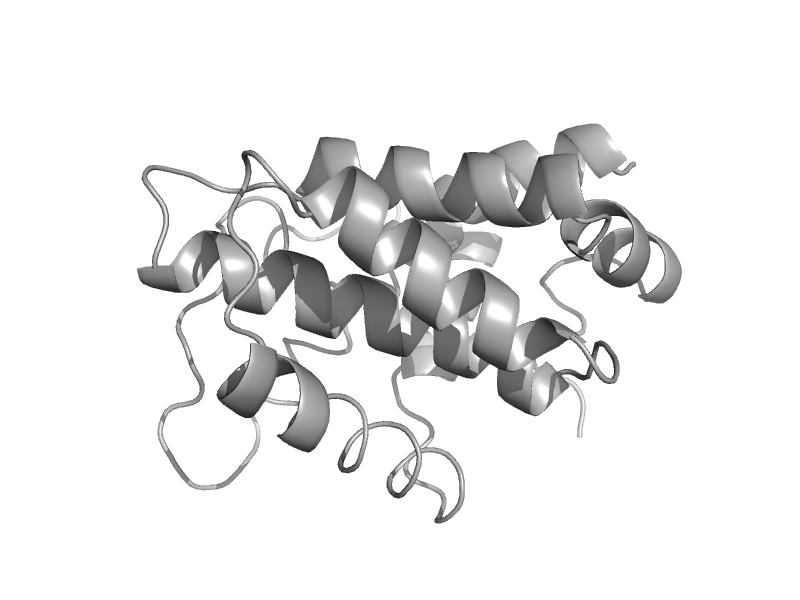 | view | |
| automated matches | automated matches | Chicken (Gallus gallus) [TaxId: 9031] |  | view | |
| automated matches | automated matches | Zebrafish (Danio rerio) [TaxId: 7955] | 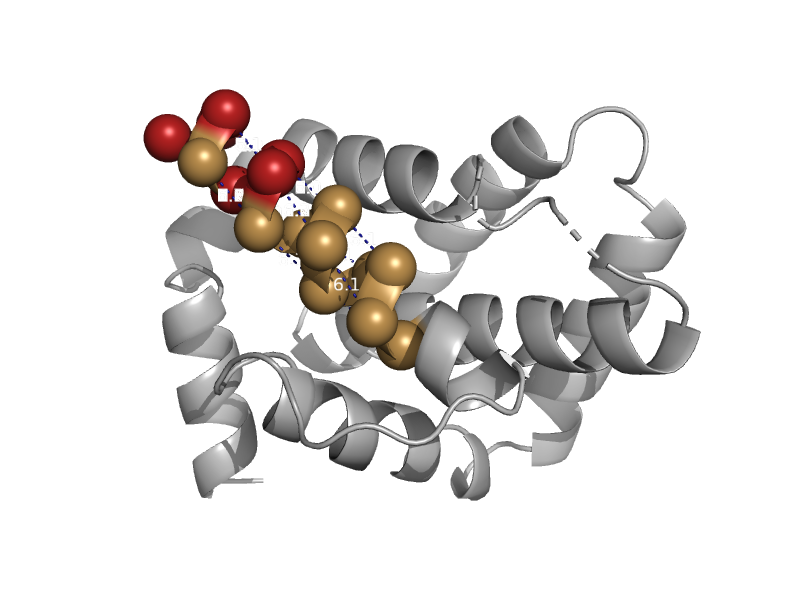 | view | |
| EAT/MCL-1 (Myeloid cell leukemia sequence 1) | Bcl-2 inhibitors of programmed cell death | Human (Homo sapiens) [TaxId: 9606] |  | view | |
| automated matches | automated matches | Trichuris suis [TaxId: 68888] | 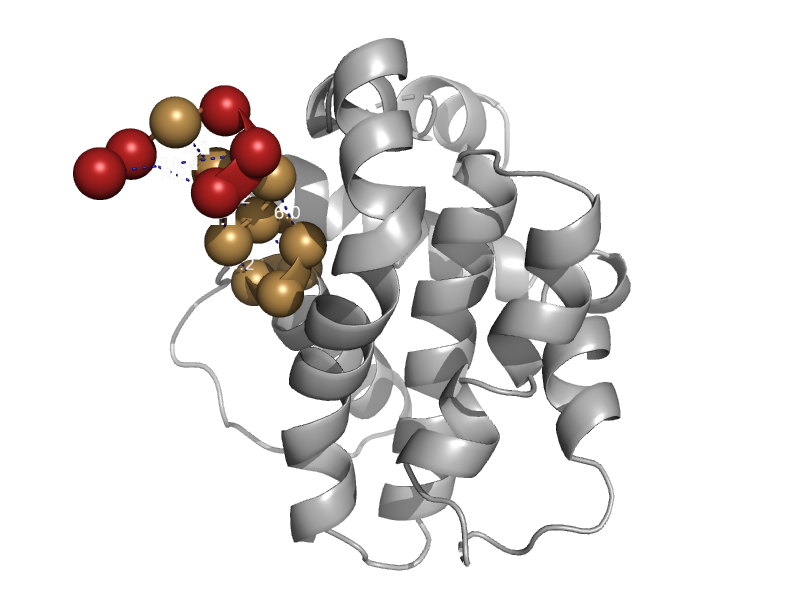 | view |
| Domain ID | Name |
|---|---|
| d7jgwa1 | 7jgw A:1-196 |
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | D | S | E | V | S | N | G | S | G | L | G | A | K | H | I | T | D | L | L | V | F | G | F | L | Q | S | S | G | C | T | R | Q | E | L | E | V | L | G | R | E | L | P | V | Q | A | Y | W | E | A | D | L | E | D | E | L | Q | T | D | G | S | Q | A | S | R | S | F | N | Q | G | R | I | E | P | D | S | E | S | Q | E | E | I | I | H | N | I | A | R | H | L | A | Q | I | G | D | E | M | D | H | N | I | Q | P | T | L | V | R | Q | L | A | A | Q | F | M | N | G | S | L | S | E | E | D | K | R | N | C | L | A | K | A | L | D | E | V | K | T | A | F | - | - | - | P | R | D | M | E | N | D | K | A | M | L | I | M | T | M | L | L | A | K | K | V | A | S | - | H | A | P | - | S | L | L | R | D | V | F | H | T | T | V | N | F | I | N | Q | N | L | F | S | Y | V | R | N | L | V | R | N | E | M | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | M | D | E | D | V | L | P | G | E | V | L | A | I | E | G | I | F | M | A | C | G | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | E | P | E | - | - | - | - | - | - | - | - | - | - | Y | L | Y | H | P | L | L | S | P | I | K | L | Y | I | T | G | L | M | R | D | K | E | - | - | - | - | S | L | F | E | A | M | L | A | N | - | - | - | - | - | - | V | R | F | H | S | T | T | G | I | D | Q | L | G | L | S | M | L | Q | V | S | G | D | G | N | M | N | W | G | R | A | L | A | I | L | T | F | G | S | F | V | A | Q | K | L | S | N | - | - | - | - | E | P | H | L | R | D | F | A | L | A | V | L | P | A | Y | A | Y | E | A | I | G | P | Q | W | F | R | A | R | G | G | W | R | - | G | L | K | A | Y | C | T | Q | V | L | T | - | - |
| - | - | - | - | - | - | D | W | E | - | E | P | R | L | D | I | E | G | F | V | V | D | Y | F | T | H | R | I | R | Q | N | G | E | W | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | A | P | G | - | L | P | S | - | - | - | - | - | - | - | - | - | - | G | V | Q | P | E | H | E | R | V | G | T | I | F | E | K | K | H | A | - | - | - | - | E | N | F | E | T | F | S | E | Q | L | L | A | - | - | - | - | V | P | I | S | F | S | L | Y | Q | D | V | V | R | T | V | G | N | - | - | - | - | - | - | P | S | Y | G | R | L | I | G | L | I | S | F | G | G | F | V | A | A | K | E | S | V | - | - | - | E | L | Q | G | Q | V | R | N | L | F | V | Y | T | S | L | F | I | K | T | R | I | - | - | N | W | R | S | W | D | D | F | T | L | - | G | Q | K | E | D | Y | E | A | E | A | - | - | - |
| - | - | - | - | - | - | M | - | - | - | - | - | A | Y | S | T | R | E | I | L | L | A | L | C | I | R | D | S | R | V | H | G | N | - | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | L | H | P | V | L | E | L | A | A | R | E | T | P | L | R | L | S | P | E | D | T | V | V | L | R | Y | H | V | L | L | E | E | I | I | E | - | - | - | - | R | N | S | E | T | F | T | E | T | W | N | R | F | I | T | H | T | E | H | V | D | L | D | F | N | S | V | F | L | E | I | F | H | R | - | - | - | G | D | P | S | L | G | R | A | L | A | W | M | A | W | C | M | H | A | C | R | T | L | C | C | N | Q | S | T | P | Y | Y | V | V | D | L | S | V | R | G | M | L | E | A | S | E | G | L | D | - | - | G | W | I | H | Q | Q | G | G | W | S | - | T | L | I | E | D | N | I | P | - | G | - | - | - |
| - | - | - | - | - | - | - | - | - | A | E | S | E | L | H | I | H | S | L | A | E | H | Y | L | Q | Y | V | L | Q | V | P | - | - | - | - | A | F | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | S | Q | A | C | R | V | L | Q | R | V | A | F | S | V | Q | E | - | - | - | V | E | N | L | K | S | Y | L | D | D | F | - | - | - | H | V | S | I | D | T | A | R | I | I | F | N | Q | V | - | E | K | E | F | E | D | G | - | - | - | I | I | N | W | G | R | I | V | T | I | F | A | F | G | G | V | L | L | K | K | L | E | - | - | - | - | - | - | - | S | A | Y | - | K | V | S | S | F | V | A | E | F | I | - | N | - | N | T | G | E | W | I | - | Q | N | G | G | W | E | D | G | F | I | K | K | F | E | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | K | S | G | T | Y | W | A | T | L | I | T | A | F | L | K | T | V | S | K | V | E | - | - | - | - | - | E | L | D | C | V | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | A | V | L | V | D | V | S | K | I | I | T | L | T | Q | E | F | R | R | - | H | Y | D | S | V | Y | R | A | D | Y | G | P | - | - | - | - | - | - | A | L | K | N | W | K | R | D | L | S | K | L | F | T | S | L | F | V | D | - | - | - | - | V | I | N | S | G | R | I | V | G | F | F | D | V | G | R | Y | V | C | E | E | V | L | C | P | G | S | W | - | T | E | D | H | E | L | L | N | D | C | M | T | H | F | F | I | E | N | N | L | M | N | H | F | P | L | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | G | G | P | T | S | S | E | Q | I | M | K | T | G | A | L | L | L | Q | G | F | I | Q | D | R | A | G | R | D | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | T | K | L | S | E | S | L | K | R | I | G | D | E | L | D | S | - | - | N | M | E | L | Q | R | M | I | A | - | - | - | - | - | - | - | - | A | V | D | T | S | P | R | V | F | F | R | V | A | A | D | M | F | S | D | G | - | - | - | N | F | N | W | G | R | V | V | A | L | F | Y | F | A | S | K | L | V | L | K | A | L | S | T | K | - | - | - | V | P | E | L | I | R | T | I | M | G | W | T | L | D | F | L | R | E | R | L | L | G | W | I | Q | D | Q | G | G | W | D | - | G | L | L | S | - | - | Y | F | G | T | P | T | W |
| - | - | - | - | - | - | - | S | E | E | Q | V | A | Q | D | T | E | E | V | F | R | S | Y | V | F | Y | R | H | Q | Q | E | Q | E | A | E | G | V | A | A | P | A | D | P | E | M | V | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | P | L | Q | P | S | S | T | M | G | Q | V | G | R | Q | L | A | I | I | G | D | D | - | I | N | R | R | Y | D | S | E | F | Q | T | M | L | Q | H | A | Q | P | T | A | E | N | A | Y | E | Y | F | T | K | I | A | T | S | L | F | E | S | - | - | - | - | G | I | N | W | G | R | V | V | A | L | L | G | F | G | Y | R | L | A | L | H | V | Y | Q | - | H | G | L | - | T | G | F | L | G | Q | V | T | R | F | V | V | D | F | M | L | H | H | S | I | A | R | W | I | A | Q | R | G | G | W | V | - | A | A | L | N | - | - | - | - | - | - | - | - | L |
| - | - | - | S | - | - | P | T | D | K | E | L | V | S | Q | A | K | A | L | C | R | D | Y | I | N | S | R | L | I | R | A | - | - | - | - | G | V | S | W | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | K | L | A | E | V | S | A | I | L | L | R | L | G | D | E | L | E | Y | - | I | R | P | N | V | Y | R | N | I | A | R | Q | L | N | I | S | L | H | S | E | T | V | V | T | D | A | F | L | A | V | A | A | Q | I | F | T | A | - | - | - | - | G | I | T | W | G | K | V | V | S | L | Y | A | V | A | A | G | L | A | V | D | C | V | R | - | H | A | Q | - | P | A | M | V | H | T | I | V | D | C | L | G | E | F | V | R | K | - | T | L | V | T | W | L | K | R | R | G | G | W | A | - | D | I | T | K | C | V | V | - | - | - | - | - | - |
| - | - | - | - | - | - | - | M | S | C | W | L | R | E | Q | T | L | L | L | A | E | D | Y | I | S | F | C | S | G | I | Q | - | - | - | - | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | P | P | S | E | S | A | E | A | M | R | Y | L | A | K | E | M | E | Q | - | Q | H | R | T | K | F | R | S | L | S | Q | E | F | L | D | - | - | T | C | G | A | D | P | S | K | C | L | Q | S | V | M | R | E | L | V | G | D | G | - | - | - | K | M | N | W | G | R | V | V | S | I | F | T | F | T | G | V | L | A | S | E | L | L | S | - | R | G | - | - | E | E | G | S | - | R | L | A | E | T | I | A | D | Y | L | G | G | - | E | K | Q | D | W | L | V | E | N | G | G | W | E | - | G | F | C | R | F | F | H | N | A | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | D | L | Y | R | Q | S | L | E | I | I | S | R | Y | L | R | E | Q | A | T | G | A | - | - | - | - | K | D | T | K | P | M | G | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | G | A | T | S | R | K | A | L | E | T | L | R | R | V | G | D | G | V | Q | R | - | N | H | E | T | A | F | Q | G | M | L | R | K | L | D | I | - | - | K | N | E | D | - | D | V | K | S | L | S | R | V | M | I | H | V | F | S | D | G | - | - | - | V | T | N | W | G | R | I | V | T | L | I | S | F | G | A | F | V | A | K | H | L | K | T | - | I | N | Q | - | E | S | C | I | E | P | L | A | E | S | I | T | D | V | L | V | R | - | T | K | R | D | W | L | V | K | Q | R | G | W | D | - | G | F | V | E | F | F | H | V | E | D | L | E | G |
| - | - | - | W | - | - | D | N | E | L | S | G | I | N | T | S | V | S | L | A | A | D | Y | V | Y | M | R | L | A | T | E | - | - | - | - | G | F | V | F | G | I | R | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | V | R | A | P | I | R | L | C | D | A | M | F | L | M | C | D | L | F | E | R | - | F | H | D | R | Y | I | A | P | L | K | N | A | C | L | G | I | S | A | D | - | - | M | D | M | R | M | F | F | S | A | L | D | S | V | F | S | S | - | - | - | - | G | I | S | W | S | R | I | V | A | M | Y | A | F | A | G | S | V | A | L | A | C | A | R | - | Q | G | R | - | R | Q | T | V | I | A | I | P | E | W | I | M | L | Y | M | R | R | - | A | I | A | P | W | I | H | A | N | G | G | W | D | - | S | F | I | F | S | Q | D | V | L | N | G | N | H |
