solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1el0a- ( 1 ) sksmqvpfsrççfsfAeqeiplrailçyrntss--içsneGliFklk--
d1f2la- ( 5 ) vt--kçnitçskmt-skipvalLihyqqNqa--sÇgkrAIILetr--
d1g2sa- ( 1 ) trgsdisktçÇfqyshkplpwtwvrsyeftsn--sÇsqrAviFttk--
d1m8aa- ( 5 ) dçÇlgytdrilhpkfIvgftrqlanegçdinAiiFhtkk-
d1nr4a- ( 2 ) rgtnvgreçÇleyfkgaiplrkLktwyqTse--dÇsrdAiVFvTv--
d1o80a- ( 1 ) vplsrtv--rçtÇisisnqpVnprslekleiipasqfçprvEIiAtMkkk
d1rjta- ( 1 ) fpmfkrgrÇlçigpgvkavkvadiEkAsimypSnnçdkieviitlken
d1zxta1 ( 5 ) vsytpnsÇÇygfqqhpPpvqiLkewypTsp--aÇpkpGViLlTk--
d2hcca- ( 1 ) h-----faadçÇtsyisqsipçslMksyfetss--eçskpGviFltk--
d2hdma- ( 2 ) gsevsdkrtçvslttqrlpçsriktytiteg----slrAviFitk--
d2mgsa- ( 8 ) lrelrçvÇlqttq-gvhpkmisnlqvfaigpqçskvevvAslkn-
d2mp1a- ( 1 ) gtnda-edççlsvtqkPipGyivrnFhyllikdgçrvpAvvFttl--
d2ra4a- ( 3 ) dalnvpstçÇftfsskkIslqrLksyviTts--rÇpqkAViFrTk--
d3gv3a- ( 7 ) yçpÇrffesh-iaranVkhLkiln-tpnçal-qivArlknn
d4hsva- ( 6 ) gdlqçlçvkttsq-vrprhitslevikagphÇptaqlIAtLkng
d4xdxa- ( 4 ) elrçqçiktyskpfhpkfikelrviesgphÇanteiivklsd-
d5coya- ( 4 ) s--sdttpçÇfayiarplprahIkeyfyTsg--kÇsnpAVVFvTr--
d5ekia- ( 7 ) dçÇlkysqrkipakvVrsyrkqepslgçsipAIlFlprkr
d5ltla- ( 9 ) ntpstçÇlkyye-vLprrlVvGyrkaln---çhlpAIIFVtr--
d7jnya1 ( 1 ) vlevyytslrçrÇvqessvfiprrfidriqilprgngçprkEiivwkkn-
333bbbbbb bbbbb
60 70 80
d1el0a- ( 46 ) rgkeaÇAldtvgwvqrHrkmlrhçpskrk
d1f2la- ( 45 ) qhrlfÇAdpkeqwVkdAmqhLdrq
d1g2sa- ( 45 ) rgkkvçthprkkwvqkyisllktp--k-------ql
d1m8aa- ( 44 ) -klsvçAnpkqtwVkyIvrllsk
d1nr4a- ( 45 ) qgraiçSdpnnkrVknAvkyLqsl
d1o80a- ( 49 ) -gekrçLnpeskaIknLlkavsems
d1rjta- ( 49 ) -kgqRçlnPkskqArliikkverknf
d1zxta1 ( 47 ) rgrqiçAdpsknwVrqlmqrLpai--a
d2hcca- ( 41 ) kgrqvçAkpsGpgvqdÇmkklkp---ysi
d2hdma- ( 43 ) rglkvçAdpqatwvrdÇvrsmdrksnt---rnnmiq
d2mgsa- ( 51 ) -gkeiçldpeapflkkviqkildggnk-------en
d2mp1a- ( 45 ) rgrqlÇAppdqpwveriiqrlqrtSakmkrrss
d2ra4a- ( 46 ) lgkeiçAdpkekwVqnymkhlg
d3gv3a- ( 46 ) -nrqvçIdp--lwIqeyleal
d4hsva- ( 49 ) --rkiçLd-----lqallykk-------iikehles
d4xdxa- ( 46 ) -grelçldpkenwvqrvvekflkraens
d5coya- ( 45 ) knrqvçAnpekkwVreyinslem---s
d5ekia- ( 47 ) sqaelçAdpkelwVqqlmqhldkt--pspq---kpa
d5ltla- ( 49 ) --nevçTnpnddwVqeyidp----------nLpllp
d7jnya1 ( 50 ) -ksivçvdpqaewiqrmmevlrkrs--sst---lpv
bbbb aaaaaaaaaa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| CC chemokine I-309 | Interleukin 8-like chemokines | Human (Homo sapiens) [TaxId: 9606] | 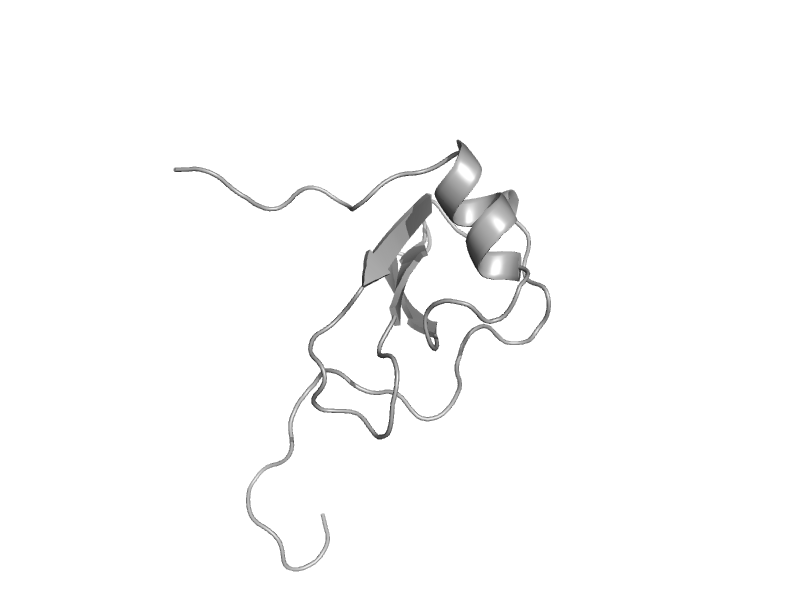 | view | |
| Chemokine domain of fractalkine | Interleukin 8-like chemokines | Human (Homo sapiens) [TaxId: 9606] |  | view | |
| Eotaxin-3 | Interleukin 8-like chemokines | Human (Homo sapiens) [TaxId: 9606] | 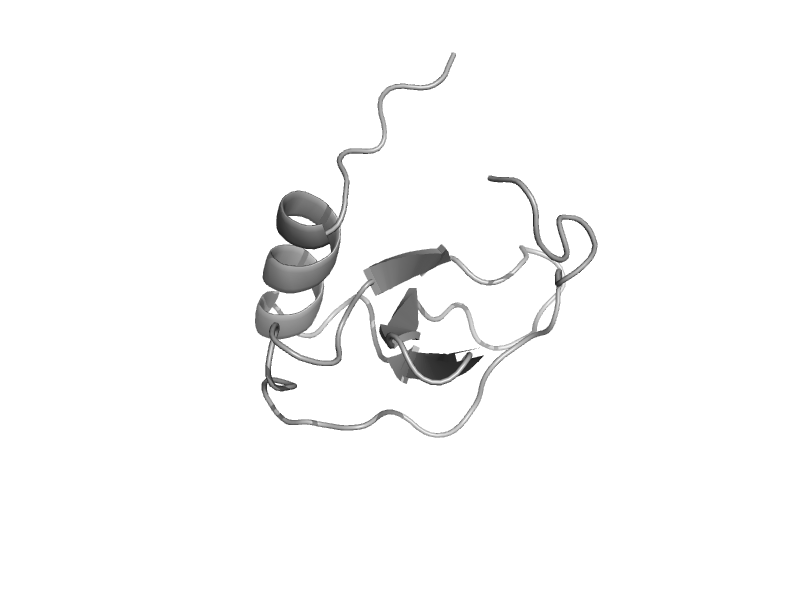 | view | |
| Macrophage inflammatory protein, MIP | Interleukin 8-like chemokines | Human (Homo sapiens), ccl20/mip-3a [TaxId: 9606] | 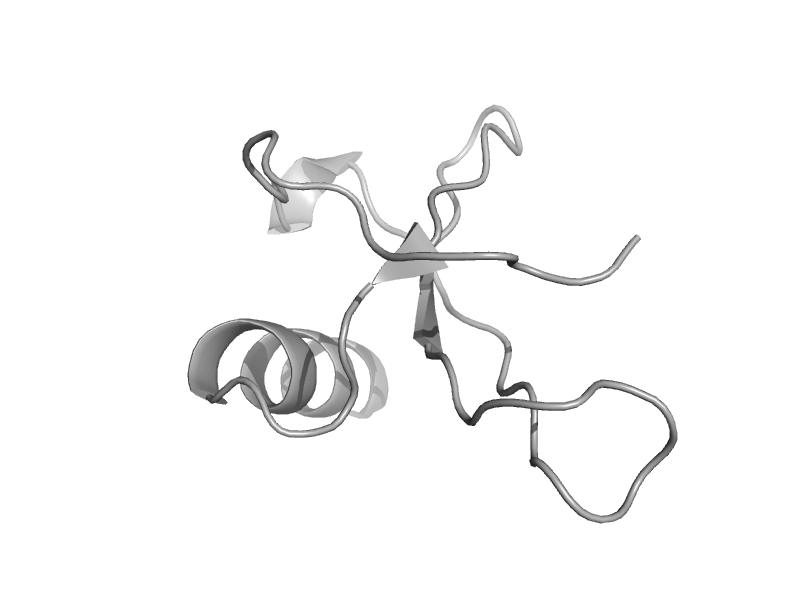 | view | |
| Thymus and activation-regulated chemokine, TRAC | Interleukin 8-like chemokines | Human (Homo sapiens) [TaxId: 9606] | 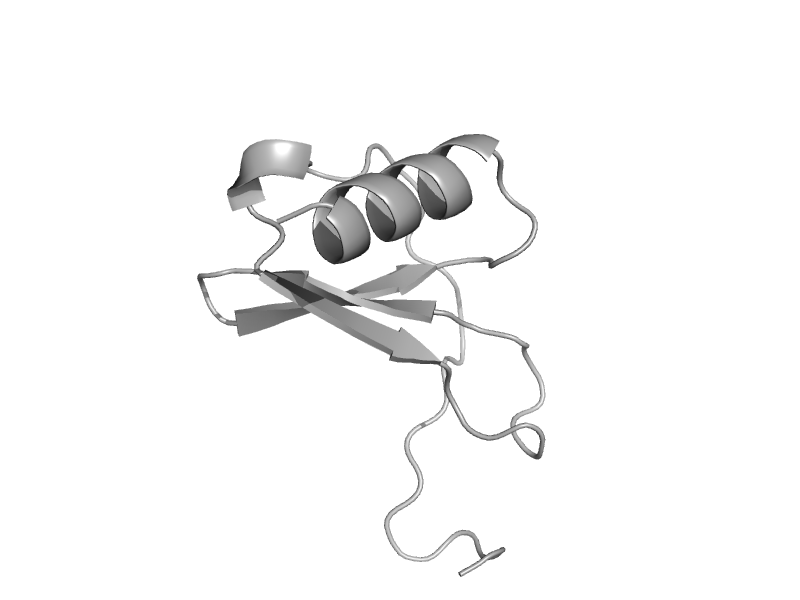 | view | |
| IP-10/CXCL10 | Interleukin 8-like chemokines | Human (Homo sapiens) [TaxId: 9606] | 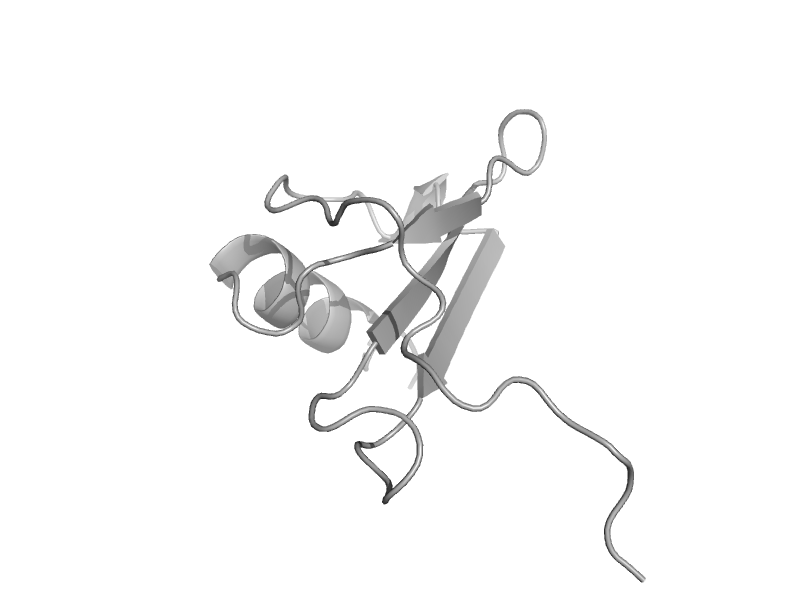 | view | |
| Small inducible cytokine b11 (CXCL11/I-TAC) | Interleukin 8-like chemokines | Human (Homo sapiens) [TaxId: 9606] |  | view | |
| automated matches | automated matches | Human herpesvirus 8 [TaxId: 37296] | 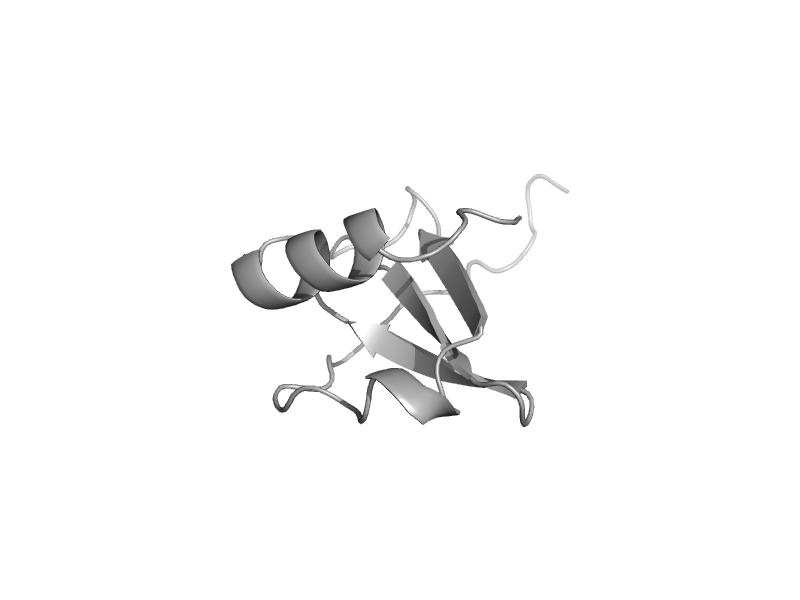 | view | |
| Chemokine hcc-2 (macrophage inflammatory protein-5) | Interleukin 8-like chemokines | Human (Homo sapiens) [TaxId: 9606] | 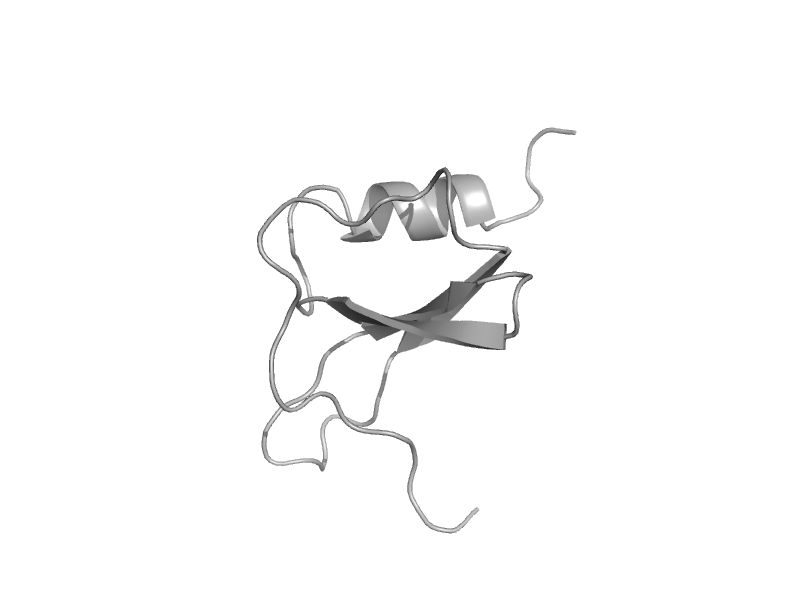 | view | |
| automated matches | Interleukin 8-like chemokines | Human (Homo sapiens) [TaxId: 9606] | 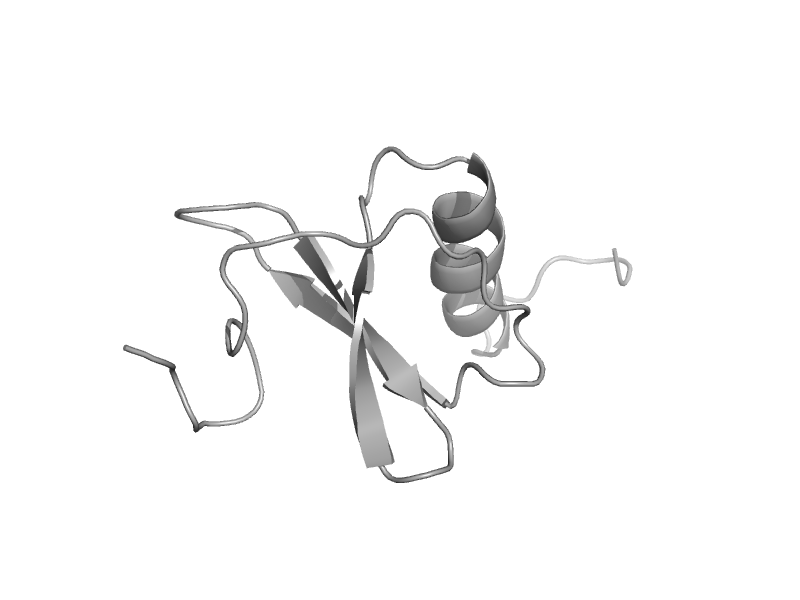 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 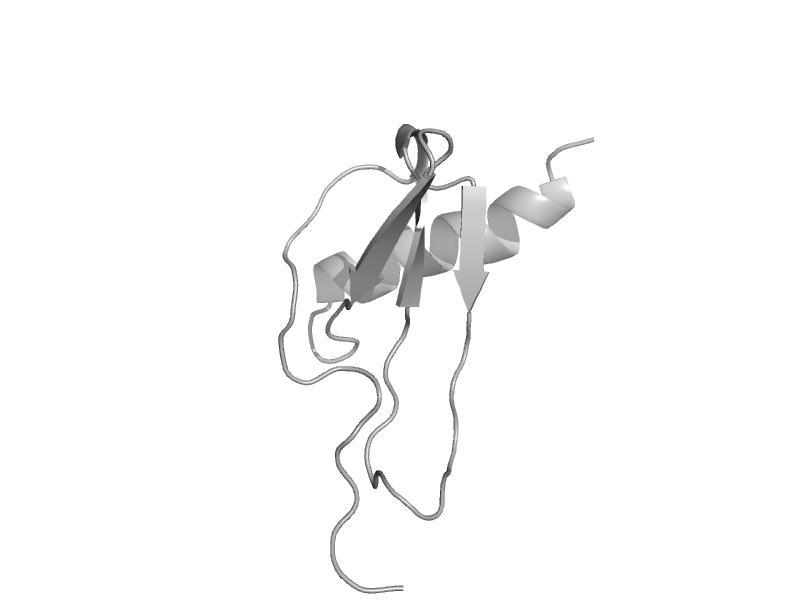 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 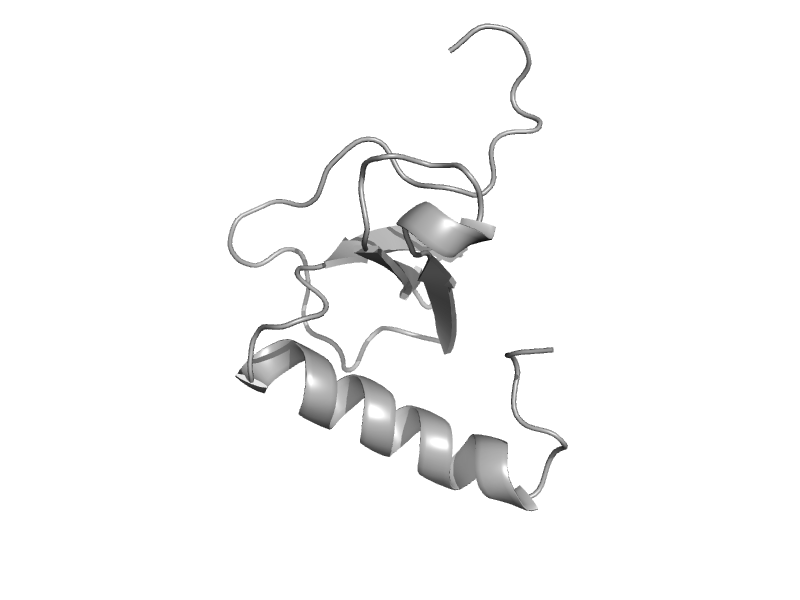 | view | |
| automated matches | Interleukin 8-like chemokines | Human (Homo sapiens) [TaxId: 9606] | 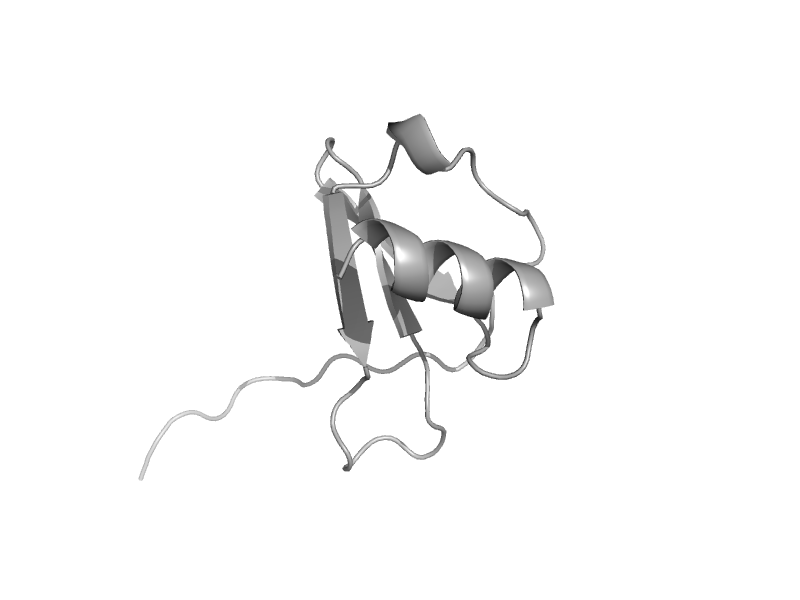 | view | |
| Stromal cell-derived factor-1 (SDF-1) | Interleukin 8-like chemokines | Human (Homo sapiens) [TaxId: 9606] | 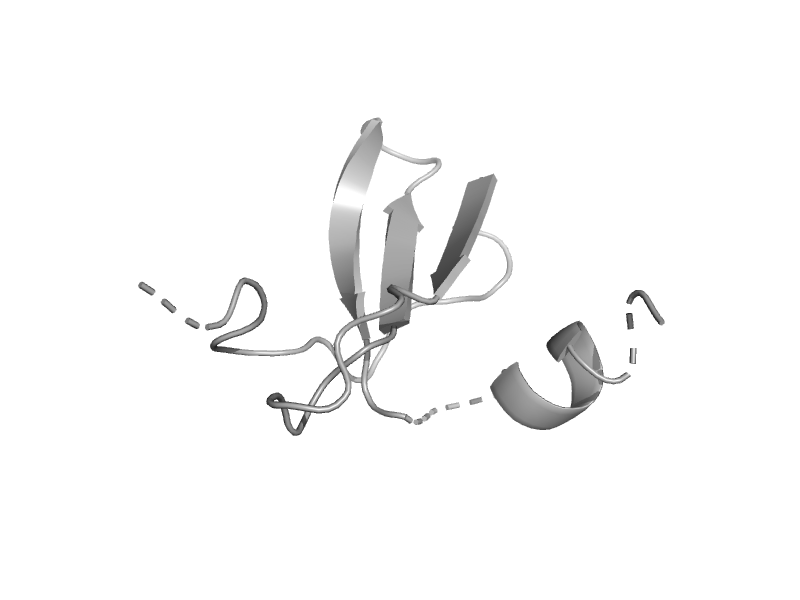 | view | |
| automated matches | Interleukin 8-like chemokines | Human (Homo sapiens) [TaxId: 9606] | 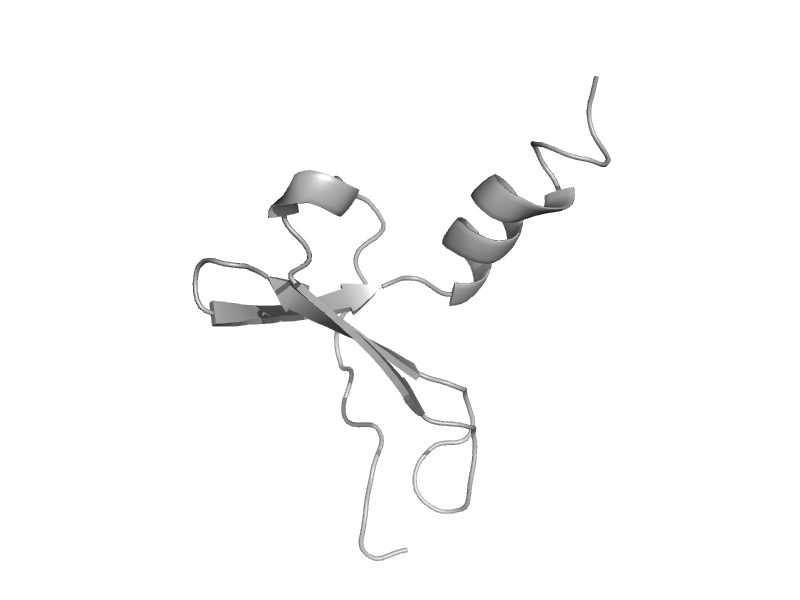 | view | |
| Interleukin-8, IL-8 | Interleukin 8-like chemokines | Human (Homo sapiens) [TaxId: 9606] | 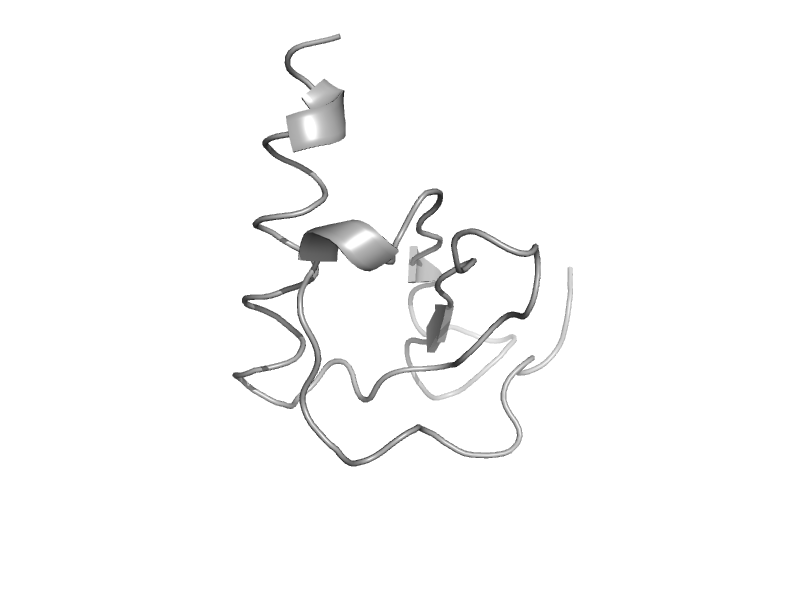 | view | |
| RANTES (regulated upon activation, normal T-cell expressed and secreted) | Interleukin 8-like chemokines | Human (Homo sapiens) [TaxId: 9606] | 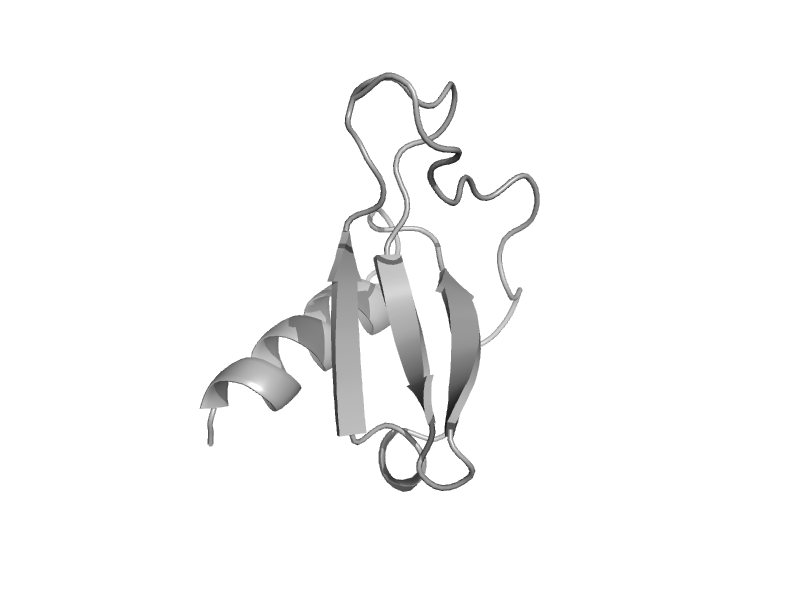 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 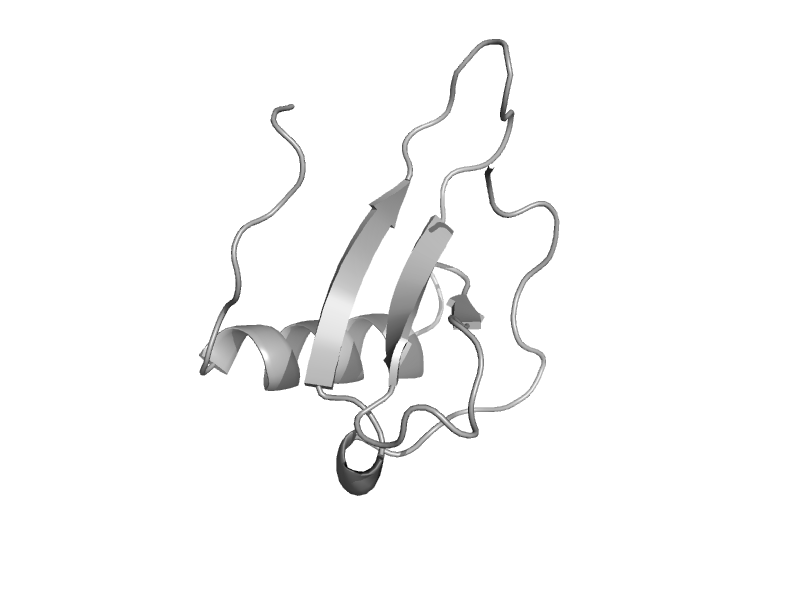 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 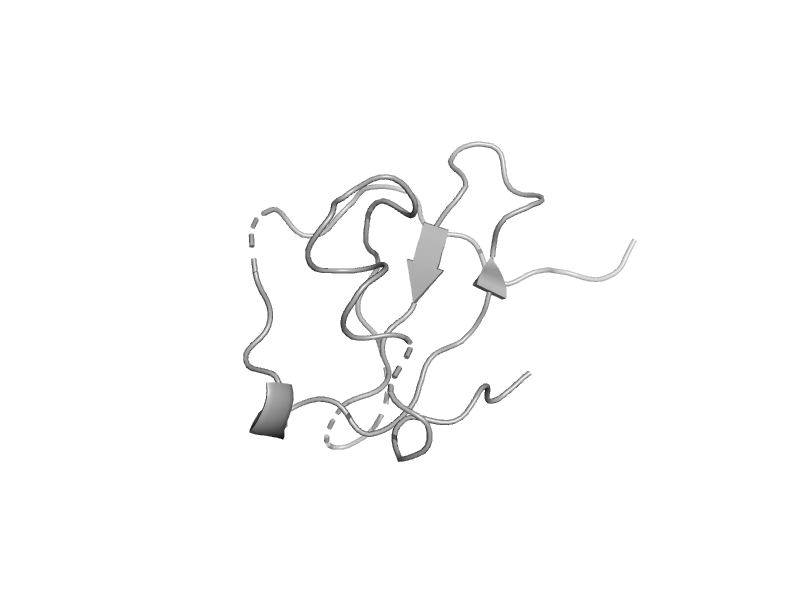 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 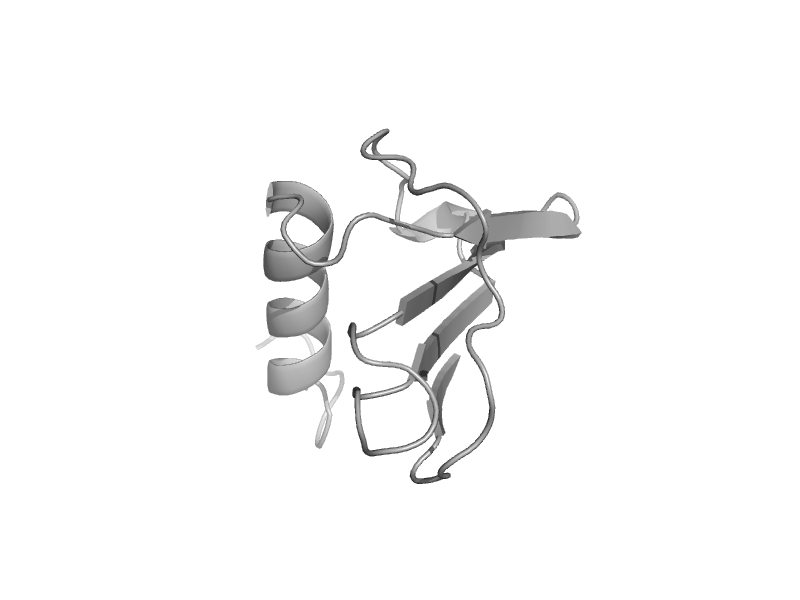 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | S | K | S | M | Q | V | P | F | S | R | C | C | F | S | F | A | E | Q | E | I | P | L | R | A | I | L | C | Y | R | N | T | S | S | - | - | I | C | S | N | E | G | L | I | F | K | L | K | - | - | R | G | K | E | A | C | A | L | D | T | V | G | W | V | Q | R | H | R | K | M | L | R | H | C | P | S | K | R | K | - | - | - | - | - | - | - |
| - | - | - | V | T | - | - | K | C | N | I | T | C | S | K | M | T | - | S | K | I | P | V | A | L | L | I | H | Y | Q | Q | N | Q | A | - | - | S | C | G | K | R | A | I | I | L | E | T | R | - | - | Q | H | R | L | F | C | A | D | P | K | E | Q | W | V | K | D | A | M | Q | H | L | D | R | Q | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | T | R | G | S | D | I | S | K | T | C | C | F | Q | Y | S | H | K | P | L | P | W | T | W | V | R | S | Y | E | F | T | S | N | - | - | S | C | S | Q | R | A | V | I | F | T | T | K | - | - | R | G | K | K | V | C | T | H | P | R | K | K | W | V | Q | K | Y | I | S | L | L | K | T | P | - | - | K | - | - | - | - | - | - | - | Q | L |
| - | - | - | - | - | - | - | - | - | - | D | C | C | L | G | Y | T | D | R | I | L | H | P | K | F | I | V | G | F | T | R | Q | L | A | N | E | G | C | D | I | N | A | I | I | F | H | T | K | K | - | - | K | L | S | V | C | A | N | P | K | Q | T | W | V | K | Y | I | V | R | L | L | S | K | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | R | G | T | N | V | G | R | E | C | C | L | E | Y | F | K | G | A | I | P | L | R | K | L | K | T | W | Y | Q | T | S | E | - | - | D | C | S | R | D | A | I | V | F | V | T | V | - | - | Q | G | R | A | I | C | S | D | P | N | N | K | R | V | K | N | A | V | K | Y | L | Q | S | L | - | - | - | - | - | - | - | - | - | - | - | - |
| V | P | L | S | R | T | V | - | - | R | C | T | C | I | S | I | S | N | Q | P | V | N | P | R | S | L | E | K | L | E | I | I | P | A | S | Q | F | C | P | R | V | E | I | I | A | T | M | K | K | K | - | G | E | K | R | C | L | N | P | E | S | K | A | I | K | N | L | L | K | A | V | S | E | M | S | - | - | - | - | - | - | - | - | - | - | - |
| - | - | F | P | M | F | K | R | G | R | C | L | C | I | G | P | G | V | K | A | V | K | V | A | D | I | E | K | A | S | I | M | Y | P | S | N | N | C | D | K | I | E | V | I | I | T | L | K | E | N | - | K | G | Q | R | C | L | N | P | K | S | K | Q | A | R | L | I | I | K | K | V | E | R | K | N | F | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | V | S | Y | T | P | N | S | C | C | Y | G | F | Q | Q | H | P | P | P | V | Q | I | L | K | E | W | Y | P | T | S | P | - | - | A | C | P | K | P | G | V | I | L | L | T | K | - | - | R | G | R | Q | I | C | A | D | P | S | K | N | W | V | R | Q | L | M | Q | R | L | P | A | I | - | - | A | - | - | - | - | - | - | - | - | - |
| - | H | - | - | - | - | - | F | A | A | D | C | C | T | S | Y | I | S | Q | S | I | P | C | S | L | M | K | S | Y | F | E | T | S | S | - | - | E | C | S | K | P | G | V | I | F | L | T | K | - | - | K | G | R | Q | V | C | A | K | P | S | G | P | G | V | Q | D | C | M | K | K | L | K | P | - | - | - | Y | S | I | - | - | - | - | - | - | - |
| - | - | - | G | S | E | V | S | D | K | R | T | C | V | S | L | T | T | Q | R | L | P | C | S | R | I | K | T | Y | T | I | T | E | G | - | - | - | - | S | L | R | A | V | I | F | I | T | K | - | - | R | G | L | K | V | C | A | D | P | Q | A | T | W | V | R | D | C | V | R | S | M | D | R | K | S | N | T | - | - | - | R | N | N | M | I | Q |
| - | - | - | - | - | L | R | E | L | R | C | V | C | L | Q | T | T | Q | - | G | V | H | P | K | M | I | S | N | L | Q | V | F | A | I | G | P | Q | C | S | K | V | E | V | V | A | S | L | K | N | - | - | G | K | E | I | C | L | D | P | E | A | P | F | L | K | K | V | I | Q | K | I | L | D | G | G | N | K | - | - | - | - | - | - | - | E | N |
| - | - | - | G | T | N | D | A | - | E | D | C | C | L | S | V | T | Q | K | P | I | P | G | Y | I | V | R | N | F | H | Y | L | L | I | K | D | G | C | R | V | P | A | V | V | F | T | T | L | - | - | R | G | R | Q | L | C | A | P | P | D | Q | P | W | V | E | R | I | I | Q | R | L | Q | R | T | S | A | K | M | K | R | R | S | S | - | - | - |
| - | - | - | D | A | L | N | V | P | S | T | C | C | F | T | F | S | S | K | K | I | S | L | Q | R | L | K | S | Y | V | I | T | T | S | - | - | R | C | P | Q | K | A | V | I | F | R | T | K | - | - | L | G | K | E | I | C | A | D | P | K | E | K | W | V | Q | N | Y | M | K | H | L | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | Y | C | P | C | R | F | F | E | S | H | - | I | A | R | A | N | V | K | H | L | K | I | L | N | - | T | P | N | C | A | L | - | Q | I | V | A | R | L | K | N | N | - | N | R | Q | V | C | I | D | P | - | - | L | W | I | Q | E | Y | L | E | A | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | G | D | L | Q | C | L | C | V | K | T | T | S | Q | - | V | R | P | R | H | I | T | S | L | E | V | I | K | A | G | P | H | C | P | T | A | Q | L | I | A | T | L | K | N | G | - | - | R | K | I | C | L | D | - | - | - | - | - | L | Q | A | L | L | Y | K | K | - | - | - | - | - | - | - | I | I | K | E | H | L | E | S |
| - | - | - | - | - | - | - | E | L | R | C | Q | C | I | K | T | Y | S | K | P | F | H | P | K | F | I | K | E | L | R | V | I | E | S | G | P | H | C | A | N | T | E | I | I | V | K | L | S | D | - | - | G | R | E | L | C | L | D | P | K | E | N | W | V | Q | R | V | V | E | K | F | L | K | R | A | E | N | S | - | - | - | - | - | - | - | - |
| - | - | - | S | - | - | S | D | T | T | P | C | C | F | A | Y | I | A | R | P | L | P | R | A | H | I | K | E | Y | F | Y | T | S | G | - | - | K | C | S | N | P | A | V | V | F | V | T | R | - | - | K | N | R | Q | V | C | A | N | P | E | K | K | W | V | R | E | Y | I | N | S | L | E | M | - | - | - | S | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | D | C | C | L | K | Y | S | Q | R | K | I | P | A | K | V | V | R | S | Y | R | K | Q | E | P | S | L | G | C | S | I | P | A | I | L | F | L | P | R | K | R | S | Q | A | E | L | C | A | D | P | K | E | L | W | V | Q | Q | L | M | Q | H | L | D | K | T | - | - | P | S | P | Q | - | - | - | K | P | A |
| - | - | - | - | - | - | N | T | P | S | T | C | C | L | K | Y | Y | E | - | V | L | P | R | R | L | V | V | G | Y | R | K | A | L | N | - | - | - | C | H | L | P | A | I | I | F | V | T | R | - | - | - | - | N | E | V | C | T | N | P | N | D | D | W | V | Q | E | Y | I | D | P | - | - | - | - | - | - | - | - | - | - | N | L | P | L | L | P |
| V | L | E | V | Y | Y | T | S | L | R | C | R | C | V | Q | E | S | S | V | F | I | P | R | R | F | I | D | R | I | Q | I | L | P | R | G | N | G | C | P | R | K | E | I | I | V | W | K | K | N | - | - | K | S | I | V | C | V | D | P | Q | A | E | W | I | Q | R | M | M | E | V | L | R | K | R | S | - | - | S | S | T | - | - | - | L | P | V |
