solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1cbfa1 ( 21 ) mkLyIIGAGPgd---pdlItvkGlklLqqAdVVLYadslVs------q
d1ve2a1 ( 1 ) mrGkVYLVGAGFGg---pehLtlkAlrVLevAevVLhd-rlVh------p
d1wdea- ( 6 ) eaVtLlLVGWgyap-----gqtleAldaVrrAdvVyVEsytpgsswL-yk
d1wyza1 ( 2 ) etALYlLpvtlgdtpleqvlpsynteIIrgIrhFIVe------dvrsAr
d2bb3a1 ( 2 ) iwIVGSGtçr----gqtterAkeiierAeVIyGs--------rrAL
d2z6ra- ( 2 ) vLyFIGLGlyd---erdItvkGleiAkkCdyVFAEfytslMagTtlg
d3ndca1 ( 1 ) MtVhFIGAGPGa---adlItirGrdlIasCpVCLyagslVp------e
d6pr1a3 ( 216 ) geVVLVGAGpgd---aglLtlkGlqqIqqAdIVVyd-rlVs------d
bbbbb aaaaaaaaa bbbb a
60 70 80 90 100
d1cbfa1 ( 60 ) dLIakSkp----gAevlkT--agmhleemvgtMldrMregkmVVrVhtgd
d1ve2a1 ( 41 ) gVlalAk------gelvpv--k-tpqeaitarLiaLAregrvVARLKgGd
d1wdea- ( 52 ) sVveaAge----arVveAs--rrdLeersreivsra-l-davVAVVTagd
d1wyza1 ( 45 ) rfLkkVdreididsltFypLnkhtspedIsgyLkplag--gasGvIse--
d2bb3a1 ( 36 ) eLAgvvdds----rAriLrsfk---gdeirr-IeeGre--reVAVIstgd
d2z6ra- ( 46 ) rIqrlIgk-----eIrvLs--redVelnFeniVLplAk-endVAFLTPgd
d3ndca1 ( 40 ) aLlahCpp----gAkivnT--apmsldaiIdtIaeAhaagqdVARLhsgd
d6pr1a3 ( 254 ) dImnlVar----dAdrvfV--g-vpqeeINqiLlreAqkgkrVVRLKggd
aaa bbbb aaaaaaa aaaa bbbbb
110 120 130 140 150
d1cbfa1 ( 104 ) PamygaimeQmvlLkregVdIeiVpGvtsVfaAaaaAeaeLtipdltqtv
d1ve2a1 ( 89 ) PmvfgrGgeEalaLrragIpfevVpGvtsAvgALsalgLpLthrglArsf
d1wdea- ( 94 ) P-vattHssLaaeAleagVaVryipGvsgvqaArga-TLsf-----yrFg
d1wyza1 ( 99 ) ----dpgadVvaiAqrqklkvipLvGPssiilSv-aSgFng-----qs-f
d2bb3a1 ( 77 ) P-vaglgrvlreia--edveikiepAiSsVqvAlarlkvdl-----se-V
d2z6ra- ( 88 ) PlvattHaelrirAkragVeSyvihApsiysaVgiT-gLhi-----ykfg
d3ndca1 ( 84 ) LsiwsamgeQlrrLralnIpydvTpGvpsfaaAaatLgaeLtlpgvaqsv
d6pr1a3 ( 304 ) PfifgrGgeEletLchagipfsvVpGitaAsGCsaySgIplthrdyAqsv
aaaaaaaaaa bbbb aaaaaaaa b
160 170 180 190 200
d1cbfa1 ( 154 ) iltrAegr---tpVpe-fekLtdlAkh----kCTIALfl-----------
d1ve2a1 ( 139 ) avatghd--palplp-----------r----adTLVLlm-----------
d1wdea- ( 139 ) gtVtLpGpwrgvtPisvArrIYlnLça----glHTTAlLdvd-ergvqLs
d1wyza1 ( 139 ) afhgyLpiepg----erakkLktLEqrVyaesqTQLFiet--------py
d2bb3a1 ( 119 ) avvdcf-----------daeltelLky-----rhLLIlAd--------sh
d2z6ra- ( 132 ) ksatVaypegnwfptsYYdvIkeNaer----glHTLLfLdikaekrmyMt
d3ndca1 ( 134 ) ilTrtsGr--asampa-getLenfArt----gaVLAIhl-----------
d6pr1a3 ( 354 ) rlvtGhlk-tggeld-----wenLAae----kQTLVFym-----------
bbbb aaaaa bbbbb
210 220 230 240 250
d1cbfa1 ( 185 ) SStlTkkVmkeFinAg-------WsedTpVVVVykAtwp-dekivrtt-V
d1ve2a1 ( 167 ) ------glkerLler--------fppetpLAlLArVgwp-geavrlgr-V
d1wdea- ( 184 ) PgqGVslLLeADreYAreagapaLLarLPSVLVe-AgaggghrVlYwssL
d1wyza1 ( 177 ) rNhkiedIlqnCrp------------qTkLCIAanItce-gefiqtrt-V
d2bb3a1 ( 149 ) --fplerLg-----------------krrVvLLEnLç-e-geriregn-A
d2z6ra- ( 178 ) AneAMelLlkVedmk----kggvFtddtlVVVLArAgsl-nptirAGy-V
d3ndca1 ( 166 ) SVhVLdeVvqkLvph--------ygedCpVAIVWraswp-dqrvvrat-L
d6pr1a3 ( 383 ) GlnqAatIqekLiafg-------mqadmpVALVEnGTsv-kQrVvhgv-L
aaaaaaaa bbbbbb bbbbbb
260 270 280 290 300
d1cbfa1 ( 226 ) kdLddaMrtngIrkqAMILAg-----wALdp
d1ve2a1 ( 201 ) edLpglge--glpspalLVVG-----kVVglYge-------------llp
d1wdea- ( 233 ) erLSta---dveggvYsIVIPArLsgvEewlLAaaSgqrrpLeydrsVye
d1wyza1 ( 214 ) kdWk----ghipkipCiFLLyk
d2bb3a1 ( 178 ) dsi------eLesdyTIIFVerev
d2z6ra- ( 222 ) kdLIre---dFgdppHILIVPgklhivEAeyLVeiAgApreilrvn----
d3ndca1 ( 206 ) atLqtslGaeler-tALILVG-----rSLatedf
d6pr1a3 ( 424 ) tqLgelAq--qVespALIIVG-----rVVaLrdk-------------lnw
bbbbb
310
d1cbfa1
d1ve2a1 ( 232 ) dhgl
d1wdea- ( 280 ) tVeeNçkkgvYepv
d1wyza1
d2bb3a1
d2z6ra- ( 265 ) -------------v
d3ndca1
d6pr1a3 ( 454 ) fsnh
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Cobalt precorrin-4 methyltransferase CbiF | Tetrapyrrole methylase | Bacillus megaterium [TaxId: 1404] | 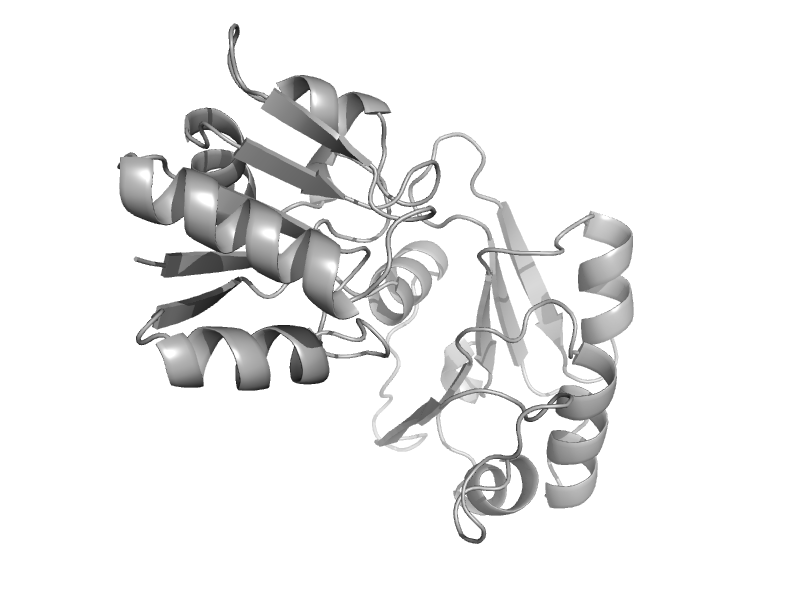 | view | |
| Uroporphyrin-III C-methyltransferase (SUMT, UROM, CobA) | Tetrapyrrole methylase | Thermus thermophilus [TaxId: 274] | 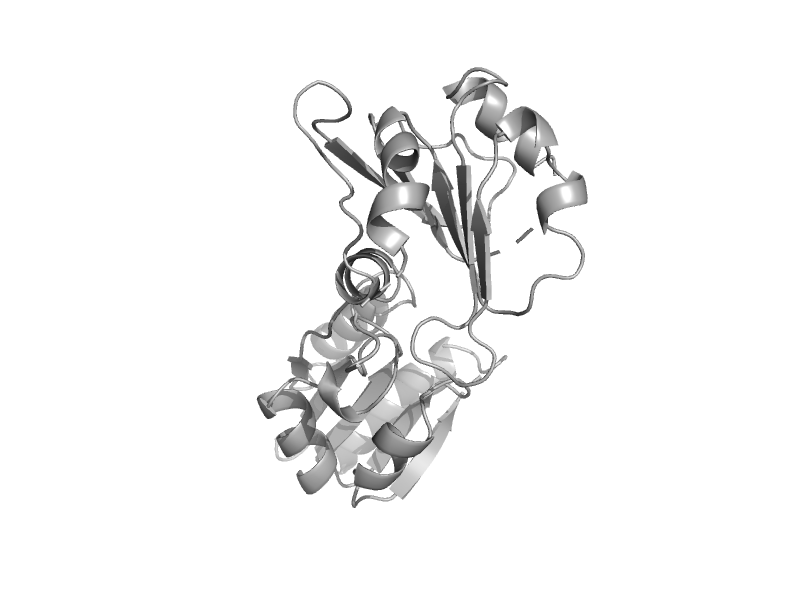 | view | |
| Diphthine synthase, DphB | Tetrapyrrole methylase | Aeropyrum pernix [TaxId: 56636] | 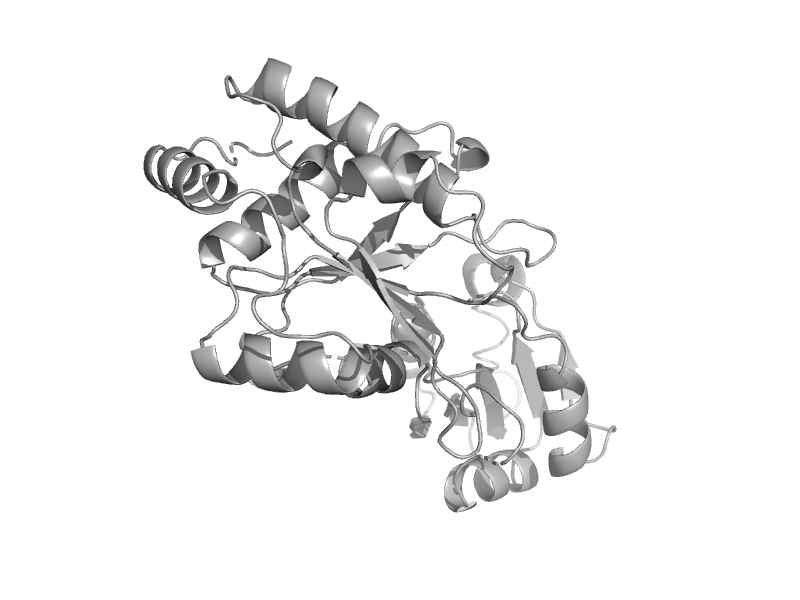 | view | |
| Putative methytransferase BT4190 | Tetrapyrrole methylase | Bacteroides thetaiotaomicron [TaxId: 818] | 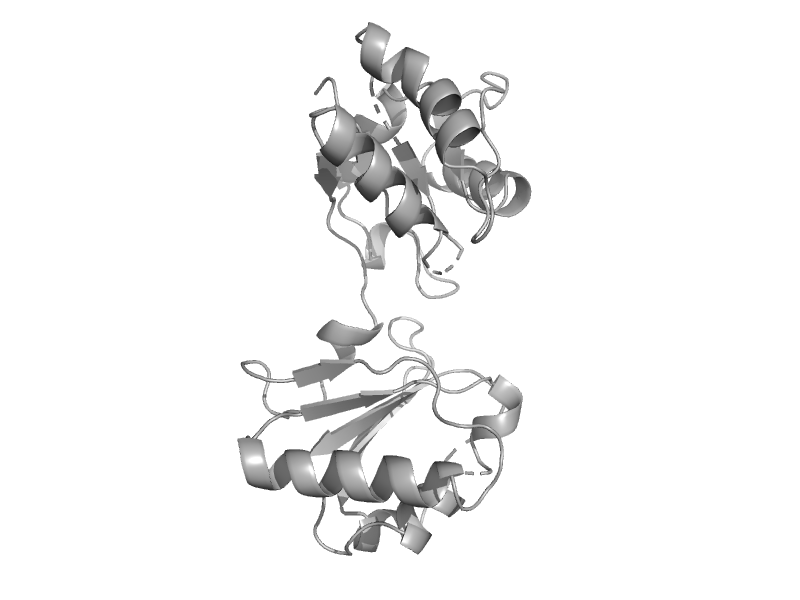 | view | |
| Precorrin-6y methylase CbiE | Tetrapyrrole methylase | Archaeoglobus fulgidus [TaxId: 2234] |  | view | |
| Diphthine synthase, DphB | Tetrapyrrole methylase | Pyrococcus horikoshii [TaxId: 53953] |  | view | |
| automated matches | automated matches | Rhodobacter capsulatus [TaxId: 272942] |  | view | |
| automated matches | Tetrapyrrole methylase | Salmonella typhimurium [TaxId: 90371] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | M | K | L | Y | I | I | G | A | G | P | G | D | - | - | - | P | D | L | I | T | V | K | G | L | K | L | L | Q | Q | A | D | V | V | L | Y | A | D | S | L | V | S | - | - | - | - | - | - | Q | D | L | I | A | K | S | K | P | - | - | - | - | G | A | E | V | L | K | T | - | - | A | G | M | H | L | E | E | M | V | G | T | M | L | D | R | M | R | E | G | K | M | V | V | R | V | H | T | G | D | P | A | M | Y | G | A | I | M | E | Q | M | V | L | L | K | R | E | G | V | D | I | E | I | V | P | G | V | T | S | V | F | A | A | A | A | A | A | E | A | E | L | T | I | P | D | L | T | Q | T | V | I | L | T | R | A | E | G | R | - | - | - | T | P | V | P | E | - | F | E | K | L | T | D | L | A | K | H | - | - | - | - | K | C | T | I | A | L | F | L | - | - | - | - | - | - | - | - | - | - | - | S | S | T | L | T | K | K | V | M | K | E | F | I | N | A | G | - | - | - | - | - | - | - | W | S | E | D | T | P | V | V | V | V | Y | K | A | T | W | P | - | D | E | K | I | V | R | T | T | - | V | K | D | L | D | D | A | M | R | T | N | G | I | R | K | Q | A | M | I | L | A | G | - | - | - | - | - | W | A | L | D | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M | R | G | K | V | Y | L | V | G | A | G | F | G | G | - | - | - | P | E | H | L | T | L | K | A | L | R | V | L | E | V | A | E | V | V | L | H | D | - | R | L | V | H | - | - | - | - | - | - | P | G | V | L | A | L | A | K | - | - | - | - | - | - | G | E | L | V | P | V | - | - | K | - | T | P | Q | E | A | I | T | A | R | L | I | A | L | A | R | E | G | R | V | V | A | R | L | K | G | G | D | P | M | V | F | G | R | G | G | E | E | A | L | A | L | R | R | A | G | I | P | F | E | V | V | P | G | V | T | S | A | V | G | A | L | S | A | L | G | L | P | L | T | H | R | G | L | A | R | S | F | A | V | A | T | G | H | D | - | - | P | A | L | P | L | P | - | - | - | - | - | - | - | - | - | - | - | R | - | - | - | - | A | D | T | L | V | L | L | M | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | L | K | E | R | L | L | E | R | - | - | - | - | - | - | - | - | F | P | P | E | T | P | L | A | L | L | A | R | V | G | W | P | - | G | E | A | V | R | L | G | R | - | V | E | D | L | P | G | L | G | E | - | - | G | L | P | S | P | A | L | L | V | V | G | - | - | - | - | - | K | V | V | G | L | Y | G | E | - | - | - | - | - | - | - | - | - | - | - | - | - | L | L | P | D | H | G | L | - | - | - | - | - | - | - | - | - | - |
| E | A | V | T | L | L | L | V | G | W | G | Y | A | P | - | - | - | - | - | G | Q | T | L | E | A | L | D | A | V | R | R | A | D | V | V | Y | V | E | S | Y | T | P | G | S | S | W | L | - | Y | K | S | V | V | E | A | A | G | E | - | - | - | - | A | R | V | V | E | A | S | - | - | R | R | D | L | E | E | R | S | R | E | I | V | S | R | A | - | L | - | D | A | V | V | A | V | V | T | A | G | D | P | - | V | A | T | T | H | S | S | L | A | A | E | A | L | E | A | G | V | A | V | R | Y | I | P | G | V | S | G | V | Q | A | A | R | G | A | - | T | L | S | F | - | - | - | - | - | Y | R | F | G | G | T | V | T | L | P | G | P | W | R | G | V | T | P | I | S | V | A | R | R | I | Y | L | N | L | C | A | - | - | - | - | G | L | H | T | T | A | L | L | D | V | D | - | E | R | G | V | Q | L | S | P | G | Q | G | V | S | L | L | L | E | A | D | R | E | Y | A | R | E | A | G | A | P | A | L | L | A | R | L | P | S | V | L | V | E | - | A | G | A | G | G | G | H | R | V | L | Y | W | S | S | L | E | R | L | S | T | A | - | - | - | D | V | E | G | G | V | Y | S | I | V | I | P | A | R | L | S | G | V | E | E | W | L | L | A | A | A | S | G | Q | R | R | P | L | E | Y | D | R | S | V | Y | E | T | V | E | E | N | C | K | K | G | V | Y | E | P | V |
| - | E | T | A | L | Y | L | L | P | V | T | L | G | D | T | P | L | E | Q | V | L | P | S | Y | N | T | E | I | I | R | G | I | R | H | F | I | V | E | - | - | - | - | - | - | D | V | R | S | A | R | R | F | L | K | K | V | D | R | E | I | D | I | D | S | L | T | F | Y | P | L | N | K | H | T | S | P | E | D | I | S | G | Y | L | K | P | L | A | G | - | - | G | A | S | G | V | I | S | E | - | - | - | - | - | - | D | P | G | A | D | V | V | A | I | A | Q | R | Q | K | L | K | V | I | P | L | V | G | P | S | S | I | I | L | S | V | - | A | S | G | F | N | G | - | - | - | - | - | Q | S | - | F | A | F | H | G | Y | L | P | I | E | P | G | - | - | - | - | E | R | A | K | K | L | K | T | L | E | Q | R | V | Y | A | E | S | Q | T | Q | L | F | I | E | T | - | - | - | - | - | - | - | - | P | Y | R | N | H | K | I | E | D | I | L | Q | N | C | R | P | - | - | - | - | - | - | - | - | - | - | - | - | Q | T | K | L | C | I | A | A | N | I | T | C | E | - | G | E | F | I | Q | T | R | T | - | V | K | D | W | K | - | - | - | - | G | H | I | P | K | I | P | C | I | F | L | L | Y | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | I | W | I | V | G | S | G | T | C | R | - | - | - | - | G | Q | T | T | E | R | A | K | E | I | I | E | R | A | E | V | I | Y | G | S | - | - | - | - | - | - | - | - | R | R | A | L | E | L | A | G | V | V | D | D | S | - | - | - | - | R | A | R | I | L | R | S | F | K | - | - | - | G | D | E | I | R | R | - | I | E | E | G | R | E | - | - | R | E | V | A | V | I | S | T | G | D | P | - | V | A | G | L | G | R | V | L | R | E | I | A | - | - | E | D | V | E | I | K | I | E | P | A | I | S | S | V | Q | V | A | L | A | R | L | K | V | D | L | - | - | - | - | - | S | E | - | V | A | V | V | D | C | F | - | - | - | - | - | - | - | - | - | - | - | D | A | E | L | T | E | L | L | K | Y | - | - | - | - | - | R | H | L | L | I | L | A | D | - | - | - | - | - | - | - | - | S | H | - | - | F | P | L | E | R | L | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | R | R | V | V | L | L | E | N | L | C | - | E | - | G | E | R | I | R | E | G | N | - | A | D | S | I | - | - | - | - | - | - | E | L | E | S | D | Y | T | I | I | F | V | E | R | E | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | V | L | Y | F | I | G | L | G | L | Y | D | - | - | - | E | R | D | I | T | V | K | G | L | E | I | A | K | K | C | D | Y | V | F | A | E | F | Y | T | S | L | M | A | G | T | T | L | G | R | I | Q | R | L | I | G | K | - | - | - | - | - | E | I | R | V | L | S | - | - | R | E | D | V | E | L | N | F | E | N | I | V | L | P | L | A | K | - | E | N | D | V | A | F | L | T | P | G | D | P | L | V | A | T | T | H | A | E | L | R | I | R | A | K | R | A | G | V | E | S | Y | V | I | H | A | P | S | I | Y | S | A | V | G | I | T | - | G | L | H | I | - | - | - | - | - | Y | K | F | G | K | S | A | T | V | A | Y | P | E | G | N | W | F | P | T | S | Y | Y | D | V | I | K | E | N | A | E | R | - | - | - | - | G | L | H | T | L | L | F | L | D | I | K | A | E | K | R | M | Y | M | T | A | N | E | A | M | E | L | L | L | K | V | E | D | M | K | - | - | - | - | K | G | G | V | F | T | D | D | T | L | V | V | V | L | A | R | A | G | S | L | - | N | P | T | I | R | A | G | Y | - | V | K | D | L | I | R | E | - | - | - | D | F | G | D | P | P | H | I | L | I | V | P | G | K | L | H | I | V | E | A | E | Y | L | V | E | I | A | G | A | P | R | E | I | L | R | V | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V |
| - | - | M | T | V | H | F | I | G | A | G | P | G | A | - | - | - | A | D | L | I | T | I | R | G | R | D | L | I | A | S | C | P | V | C | L | Y | A | G | S | L | V | P | - | - | - | - | - | - | E | A | L | L | A | H | C | P | P | - | - | - | - | G | A | K | I | V | N | T | - | - | A | P | M | S | L | D | A | I | I | D | T | I | A | E | A | H | A | A | G | Q | D | V | A | R | L | H | S | G | D | L | S | I | W | S | A | M | G | E | Q | L | R | R | L | R | A | L | N | I | P | Y | D | V | T | P | G | V | P | S | F | A | A | A | A | A | T | L | G | A | E | L | T | L | P | G | V | A | Q | S | V | I | L | T | R | T | S | G | R | - | - | A | S | A | M | P | A | - | G | E | T | L | E | N | F | A | R | T | - | - | - | - | G | A | V | L | A | I | H | L | - | - | - | - | - | - | - | - | - | - | - | S | V | H | V | L | D | E | V | V | Q | K | L | V | P | H | - | - | - | - | - | - | - | - | Y | G | E | D | C | P | V | A | I | V | W | R | A | S | W | P | - | D | Q | R | V | V | R | A | T | - | L | A | T | L | Q | T | S | L | G | A | E | L | E | R | - | T | A | L | I | L | V | G | - | - | - | - | - | R | S | L | A | T | E | D | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | G | E | V | V | L | V | G | A | G | P | G | D | - | - | - | A | G | L | L | T | L | K | G | L | Q | Q | I | Q | Q | A | D | I | V | V | Y | D | - | R | L | V | S | - | - | - | - | - | - | D | D | I | M | N | L | V | A | R | - | - | - | - | D | A | D | R | V | F | V | - | - | G | - | V | P | Q | E | E | I | N | Q | I | L | L | R | E | A | Q | K | G | K | R | V | V | R | L | K | G | G | D | P | F | I | F | G | R | G | G | E | E | L | E | T | L | C | H | A | G | I | P | F | S | V | V | P | G | I | T | A | A | S | G | C | S | A | Y | S | G | I | P | L | T | H | R | D | Y | A | Q | S | V | R | L | V | T | G | H | L | K | - | T | G | G | E | L | D | - | - | - | - | - | W | E | N | L | A | A | E | - | - | - | - | K | Q | T | L | V | F | Y | M | - | - | - | - | - | - | - | - | - | - | - | G | L | N | Q | A | A | T | I | Q | E | K | L | I | A | F | G | - | - | - | - | - | - | - | M | Q | A | D | M | P | V | A | L | V | E | N | G | T | S | V | - | K | Q | R | V | V | H | G | V | - | L | T | Q | L | G | E | L | A | Q | - | - | Q | V | E | S | P | A | L | I | I | V | G | - | - | - | - | - | R | V | V | A | L | R | D | K | - | - | - | - | - | - | - | - | - | - | - | - | - | L | N | W | F | S | N | H | - | - | - | - | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1ve2a1 | A | Q53WA2 | GO:0004851 |
| d1ve2a1 | A | Q53WA2 | GO:0006779 |
| d1ve2a1 | A | Q53WA2 | GO:0008168 |
| d1ve2a1 | A | Q53WA2 | GO:0019354 |
| d1ve2a1 | A | Q53WA2 | GO:0032259 |
| d1ve2a1 | A | Q53WA2 | GO:0004851 |
| d1ve2a1 | A | Q53WA2 | GO:0006779 |
| d1ve2a1 | A | Q53WA2 | GO:0008168 |
| d1ve2a1 | A | Q53WA2 | GO:0019354 |
| d1ve2a1 | A | Q53WA2 | GO:0032259 |
| d1wdea_ | A | Q9YDI2 | GO:0004164 |
| d1wdea_ | A | Q9YDI2 | GO:0008168 |
| d1wdea_ | A | Q9YDI2 | GO:0017183 |
| d1wdea_ | A | Q9YDI2 | GO:0032259 |
| d1wdea_ | A | Q9YDI2 | GO:0004164 |
| d1wdea_ | A | Q9YDI2 | GO:0008168 |
| d1wdea_ | A | Q9YDI2 | GO:0017183 |
| d1wdea_ | A | Q9YDI2 | GO:0032259 |
| d1wyza1 | A | Q8A031 | GO:0008168 |
| d1wyza1 | A | Q8A031 | GO:0008168 |
| d2bb3a1 | A | O29536 | GO:0008168 |
| d2bb3a1 | A | O29536 | GO:0008276 |
| d2bb3a1 | A | O29536 | GO:0009236 |
| d2bb3a1 | A | O29536 | GO:0032259 |
| d2bb3a1 | A | O29536 | GO:0008168 |
| d2bb3a1 | A | O29536 | GO:0008276 |
| d2bb3a1 | A | O29536 | GO:0009236 |
| d2bb3a1 | A | O29536 | GO:0032259 |
