solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1m3sa1 ( 1 ) mtteyvaeilnelhnSaa--yis-----
d1nria- ( 9 ) lstliTeqrNpnSvdidrqstleivrlneedklVplaies---Cl-----
d1vima1 ( 2 ) lrflevvseHiknlrn--hid-----
d1wiwa- ( 2 ) rdLdreetylvDrtgLaleLrdLvgtgpvPg
d2i2wa- ( 1 ) myqdlirnelneaaeTlanFlkddani-----
d2yvaa- ( 1 ) mqerikacftesiqtqiaAae--alp-----
d5lu5a- ( 3 ) nreltyitnsiaeaqrVmaaMladerll-----
aaaaaaaaaaaaaaaa
60 70 80 90 100
d1m3sa1 ( 23 ) ------------neadqLAdhIlsSh------qIFTAGag----------
d1nria- ( 52 ) ------------pqislAveqIvqafqq--gGrLIYIGag----------
d1vima1 ( 21 ) ------------letvgeMilIdsar------SIFViGag----------
d1wiwa- ( 33 ) eaYpgphAAlGygEGqfAAlLSglpdwgeegTLFlLEGGydlgeaAgaae
d2i2wa- ( 28 ) ------------haIqrAAvlLAdsFka--ggkVLSCGng----------
d2yvaa- ( 25 ) ------------daisrAamtLvqsLln--gnkILCCGng----------
d5lu5a- ( 31 ) ------------atvrkVAdaCiaSiaq--ggkVLLAGng----------
aaaaaaaaaaaa bbbb
110 120 130 140 150
d1m3sa1 ( 46 ) ---------------------rsglmAksFAmrLmhMg------fnAhiv
d1nria- ( 78 ) ---------------------tsGrlGvldaseCpptfgvs-tevkGiIA
d1vima1 ( 44 ) ---------------------rsgyiAkaFamrLmHLg------ytVyvv
d1wiwa- ( 86 ) tgrArvVrVGfrpgVevhIppspLApYrYlrFLLlATgreevlrs----V
d2i2wa- ( 54 ) ---------------------gShcDAmhFAeeLtgryrenrpgyPAiaI
d2yvaa- ( 51 ) ---------------------tSaaNAqhFaaSMinrfeteRpslpAiAL
d5lu5a- ( 57 ) ---------------------gSaadAqhIagqFvsrfafdRpgLpAvAL
aaaaaaaaaaaaaaa bbb
160 170 180 190 200
d1m3sa1 ( 69 ) gei----------------------ltp-plaegDLVIIGsgsGetksli
d1nria- ( 107 ) ggecAirhp--vegaedntkaVlndLqsihFsknDVLVGIaasgrtpYVi
d1vima1 ( 67 ) get----------------------vtp-ritdqDVLVGISgsGettsVv
d1wiwa- ( 132 ) deALleErrrLgpevpveeNpAKflAytLl---erlPLFYSPl--frpLE
d2i2wa- ( 83 ) sdvshiscv--g-ndfGfndiFsryVea-vGregDVLLGIStsGnsaNVi
d2yvaa- ( 80 ) Ntdnvvlta--iandrlhdevYAkqVra-lGhagDVLLAIstrGnsrdIv
d5lu5a- ( 86 ) Ttdtsilta--IgndygyekLFsrqVqa-lGnegDVLIGYstsGkspNIl
aaaaaa bbbbb aaaa
210 220 230 240 250
d1m3sa1 ( 96 ) htAakAkslhGiVAALTinpeSsIGk---------qA---dLiIrMPGsp
d1nria- ( 155 ) aGLqyAkslgAlTIsIASNpkse-ae---------iA---diaIeTiVgp
d1vima1 ( 94 ) niSkkAkdigSkLVAVTgkrdSsLAk---------mA---dvvMvVkgkm
d1wiwa- ( 177 ) gavqtLFarVAkslSltPppsAlefFlvglearheqGdplAAVLLGpgee
d2i2wa- ( 129 ) kAIaaArekgMkVITLTGkdGgkMag---------tA---dieIrVph--
d2yvaa- ( 127 ) kAVeAAvtrdMtIVALTGydGgeLag---------lLgpqdVeIrIps--
d5lu5a- ( 133 ) aAFreAkakgMtCVGFTGnrGgeMre---------lC---dllLeVps--
aaaaaaaa bbbbbb aa bbbb
260 270 280 290 300
d1m3sa1 ( 134 ) ----kdyktiqpm----------gsLfeqTLllfYdaVilkLmekkglds
d1nria- ( 193 ) Eil------tgss----------rlKSGtAQk-VLn-LTtaSillgkcye
d1vima1 ( 133 ) qerdeilsqlApl----------gtMFelTAmiFLdaLvaeImmqkhlt-
d1wiwa- ( 227 ) aalAkeiLesrVdalaevpAtganrLAqVALwYrAWTAYYLALLyGvdPg
d2i2wa- ( 165 ) fgy--------------------adrIqeIHikVIhiLIqlIekeMvk
d2yvaa- ( 166 ) hr---------------------sarIqeMHmlTVncLCdlIDntlfph
d5lu5a- ( 169 ) ad---------------------tpkIqeGHlvLGhiVCglVEhsifg
aaaaaaaaaaaaaaaaaaaaa
310 320
d1m3sa1 ( 175 ) etmfthhanle
d1nria- ( 228 ) nlvdvqasneklkaravrivqatdcnk
d1vima1 ( 172 ) edlearhavle
d1wiwa- ( 279 ) dhglle
d2i2wa-
d2yvaa-
d5lu5a-
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Hypothetical protein YckF | mono-SIS domain | Bacillus subtilis [TaxId: 1423] | 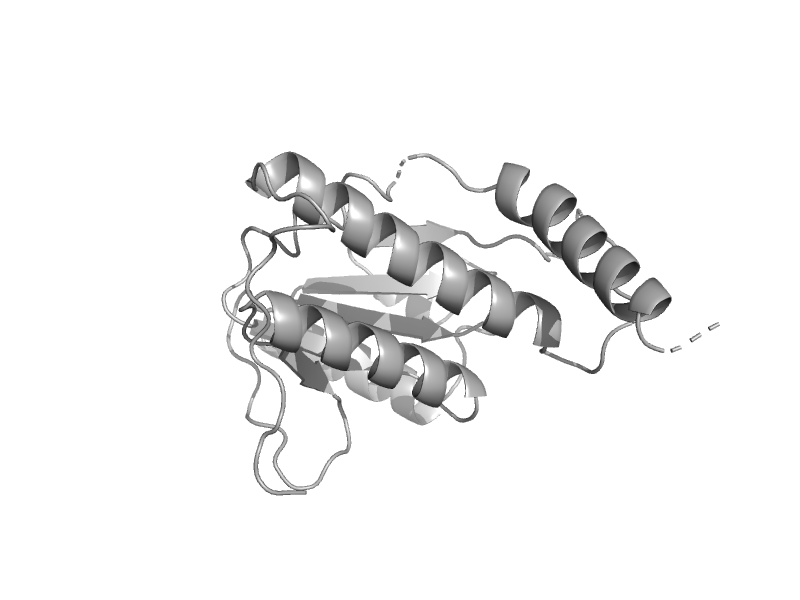 | view | |
| Hypothetical protein HI0754 | mono-SIS domain | Haemophilus influenzae [TaxId: 727] | 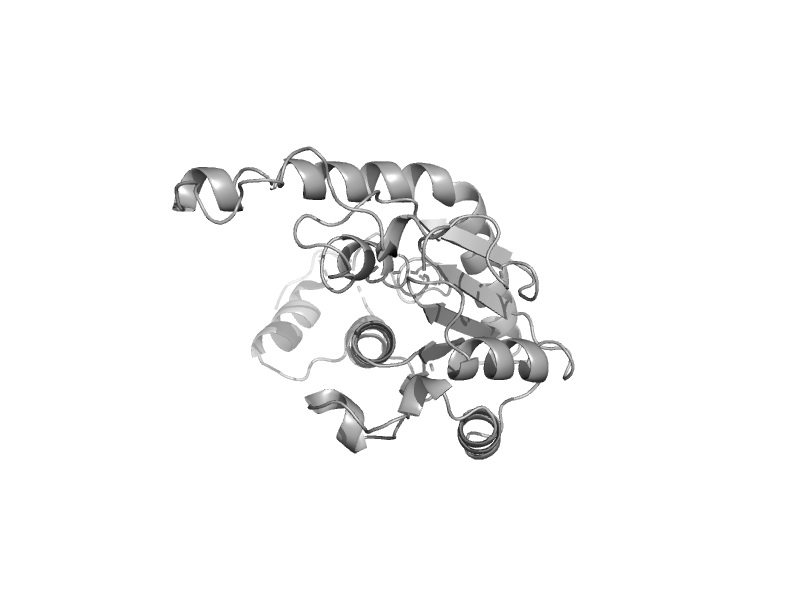 | view | |
| Hypothetical protein AF1796 | mono-SIS domain | Archaeoglobus fulgidus [TaxId: 2234] | 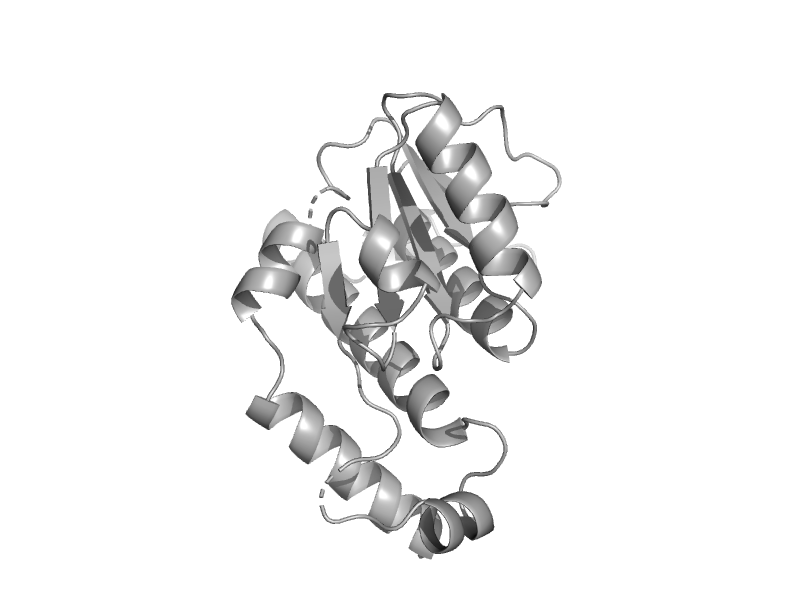 | view | |
| Glucose-6-phosphate isomerase-like protein TTHA1346 | double-SIS domain | Thermus thermophilus [TaxId: 274] | 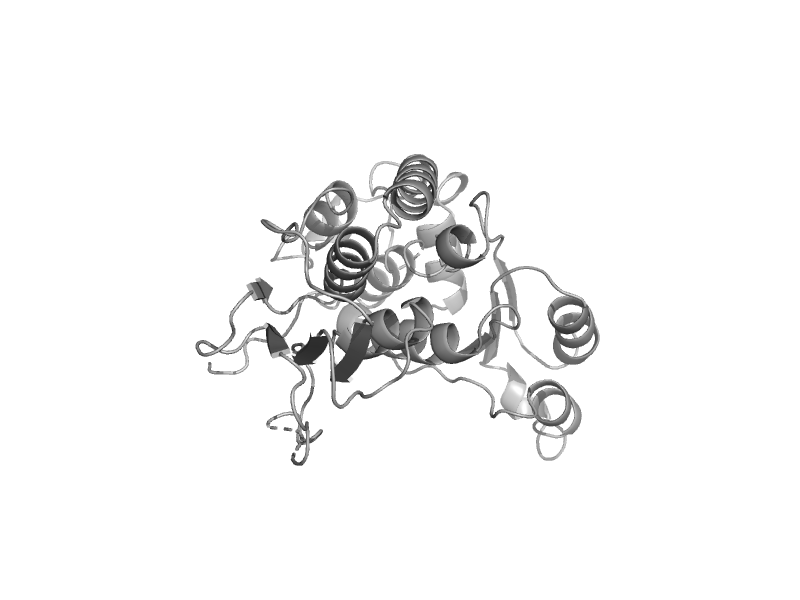 | view | |
| automated matches | automated matches | Escherichia coli [TaxId: 562] | 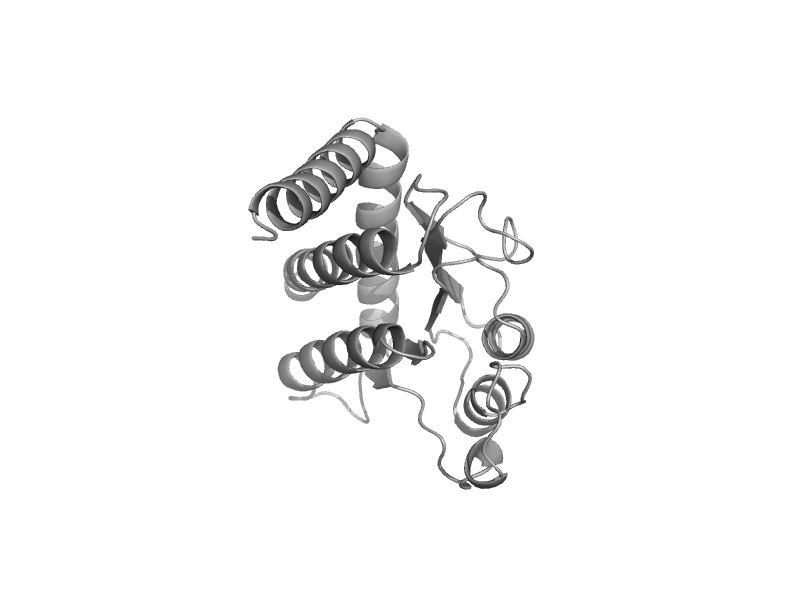 | view | |
| automated matches | mono-SIS domain | Escherichia coli [TaxId: 562] | 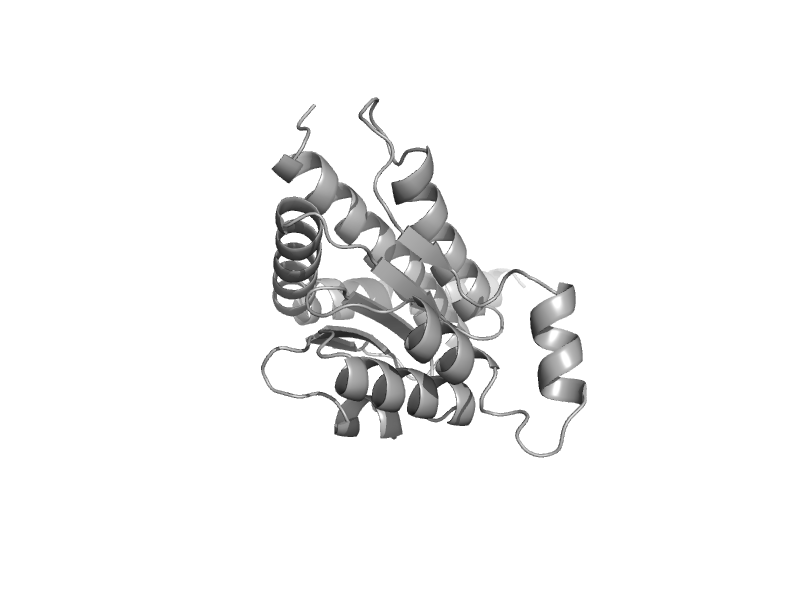 | view | |
| automated matches | automated matches | Burkholderia pseudomallei [TaxId: 272560] | 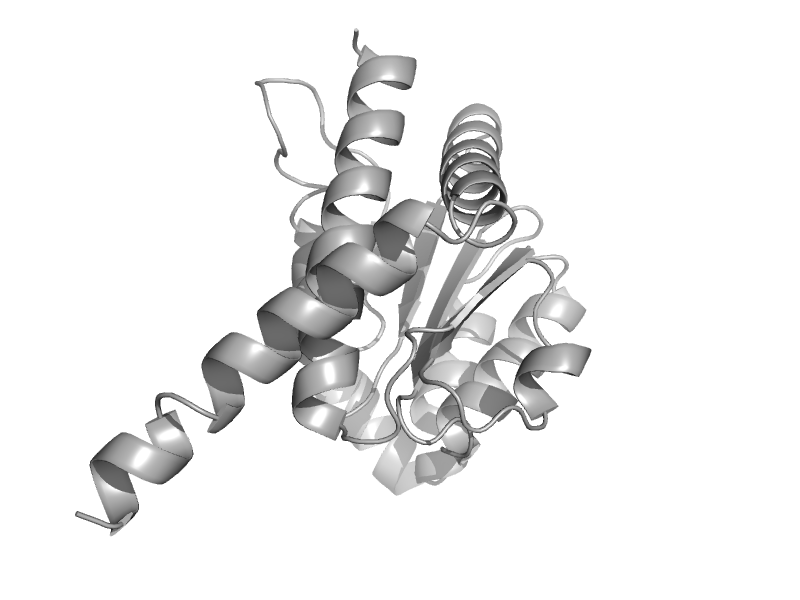 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | T | T | E | Y | V | A | E | I | L | N | E | L | H | N | S | A | A | - | - | Y | I | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | E | A | D | Q | L | A | D | H | I | L | S | S | H | - | - | - | - | - | - | Q | I | F | T | A | G | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | S | G | L | M | A | K | S | F | A | M | R | L | M | H | M | G | - | - | - | - | - | - | F | N | A | H | I | V | G | E | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | T | P | - | P | L | A | E | G | D | L | V | I | I | G | S | G | S | G | E | T | K | S | L | I | H | T | A | A | K | A | K | S | L | H | G | I | V | A | A | L | T | I | N | P | E | S | S | I | G | K | - | - | - | - | - | - | - | - | - | Q | A | - | - | - | D | L | I | I | R | M | P | G | S | P | - | - | - | - | K | D | Y | K | T | I | Q | P | M | - | - | - | - | - | - | - | - | - | - | G | S | L | F | E | Q | T | L | L | L | F | Y | D | A | V | I | L | K | L | M | E | K | K | G | L | D | S | E | T | M | F | T | H | H | A | N | L | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| L | S | T | L | I | T | E | Q | R | N | P | N | S | V | D | I | D | R | Q | S | T | L | E | I | V | R | L | N | E | E | D | K | L | V | P | L | A | I | E | S | - | - | - | C | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | Q | I | S | L | A | V | E | Q | I | V | Q | A | F | Q | Q | - | - | G | G | R | L | I | Y | I | G | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | S | G | R | L | G | V | L | D | A | S | E | C | P | P | T | F | G | V | S | - | T | E | V | K | G | I | I | A | G | G | E | C | A | I | R | H | P | - | - | V | E | G | A | E | D | N | T | K | A | V | L | N | D | L | Q | S | I | H | F | S | K | N | D | V | L | V | G | I | A | A | S | G | R | T | P | Y | V | I | A | G | L | Q | Y | A | K | S | L | G | A | L | T | I | S | I | A | S | N | P | K | S | E | - | A | E | - | - | - | - | - | - | - | - | - | I | A | - | - | - | D | I | A | I | E | T | I | V | G | P | E | I | L | - | - | - | - | - | - | T | G | S | S | - | - | - | - | - | - | - | - | - | - | R | L | K | S | G | T | A | Q | K | - | V | L | N | - | L | T | T | A | S | I | L | L | G | K | C | Y | E | N | L | V | D | V | Q | A | S | N | E | K | L | K | A | R | A | V | R | I | V | Q | A | T | D | C | N | K |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | R | F | L | E | V | V | S | E | H | I | K | N | L | R | N | - | - | H | I | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | E | T | V | G | E | M | I | L | I | D | S | A | R | - | - | - | - | - | - | S | I | F | V | I | G | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | S | G | Y | I | A | K | A | F | A | M | R | L | M | H | L | G | - | - | - | - | - | - | Y | T | V | Y | V | V | G | E | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V | T | P | - | R | I | T | D | Q | D | V | L | V | G | I | S | G | S | G | E | T | T | S | V | V | N | I | S | K | K | A | K | D | I | G | S | K | L | V | A | V | T | G | K | R | D | S | S | L | A | K | - | - | - | - | - | - | - | - | - | M | A | - | - | - | D | V | V | M | V | V | K | G | K | M | Q | E | R | D | E | I | L | S | Q | L | A | P | L | - | - | - | - | - | - | - | - | - | - | G | T | M | F | E | L | T | A | M | I | F | L | D | A | L | V | A | E | I | M | M | Q | K | H | L | T | - | E | D | L | E | A | R | H | A | V | L | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | D | L | D | R | E | E | T | Y | L | V | D | R | T | G | L | A | L | E | L | R | D | L | V | G | T | G | P | V | P | G | E | A | Y | P | G | P | H | A | A | L | G | Y | G | E | G | Q | F | A | A | L | L | S | G | L | P | D | W | G | E | E | G | T | L | F | L | L | E | G | G | Y | D | L | G | E | A | A | G | A | A | E | T | G | R | A | R | V | V | R | V | G | F | R | P | G | V | E | V | H | I | P | P | S | P | L | A | P | Y | R | Y | L | R | F | L | L | L | A | T | G | R | E | E | V | L | R | S | - | - | - | - | V | D | E | A | L | L | E | E | R | R | R | L | G | P | E | V | P | V | E | E | N | P | A | K | F | L | A | Y | T | L | L | - | - | - | E | R | L | P | L | F | Y | S | P | L | - | - | F | R | P | L | E | G | A | V | Q | T | L | F | A | R | V | A | K | S | L | S | L | T | P | P | P | S | A | L | E | F | F | L | V | G | L | E | A | R | H | E | Q | G | D | P | L | A | A | V | L | L | G | P | G | E | E | A | A | L | A | K | E | I | L | E | S | R | V | D | A | L | A | E | V | P | A | T | G | A | N | R | L | A | Q | V | A | L | W | Y | R | A | W | T | A | Y | Y | L | A | L | L | Y | G | V | D | P | G | D | H | G | L | L | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | Y | Q | D | L | I | R | N | E | L | N | E | A | A | E | T | L | A | N | F | L | K | D | D | A | N | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | H | A | I | Q | R | A | A | V | L | L | A | D | S | F | K | A | - | - | G | G | K | V | L | S | C | G | N | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | S | H | C | D | A | M | H | F | A | E | E | L | T | G | R | Y | R | E | N | R | P | G | Y | P | A | I | A | I | S | D | V | S | H | I | S | C | V | - | - | G | - | N | D | F | G | F | N | D | I | F | S | R | Y | V | E | A | - | V | G | R | E | G | D | V | L | L | G | I | S | T | S | G | N | S | A | N | V | I | K | A | I | A | A | A | R | E | K | G | M | K | V | I | T | L | T | G | K | D | G | G | K | M | A | G | - | - | - | - | - | - | - | - | - | T | A | - | - | - | D | I | E | I | R | V | P | H | - | - | F | G | Y | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | D | R | I | Q | E | I | H | I | K | V | I | H | I | L | I | Q | L | I | E | K | E | M | V | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | Q | E | R | I | K | A | C | F | T | E | S | I | Q | T | Q | I | A | A | A | E | - | - | A | L | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | A | I | S | R | A | A | M | T | L | V | Q | S | L | L | N | - | - | G | N | K | I | L | C | C | G | N | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | S | A | A | N | A | Q | H | F | A | A | S | M | I | N | R | F | E | T | E | R | P | S | L | P | A | I | A | L | N | T | D | N | V | V | L | T | A | - | - | I | A | N | D | R | L | H | D | E | V | Y | A | K | Q | V | R | A | - | L | G | H | A | G | D | V | L | L | A | I | S | T | R | G | N | S | R | D | I | V | K | A | V | E | A | A | V | T | R | D | M | T | I | V | A | L | T | G | Y | D | G | G | E | L | A | G | - | - | - | - | - | - | - | - | - | L | L | G | P | Q | D | V | E | I | R | I | P | S | - | - | H | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | A | R | I | Q | E | M | H | M | L | T | V | N | C | L | C | D | L | I | D | N | T | L | F | P | H | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | R | E | L | T | Y | I | T | N | S | I | A | E | A | Q | R | V | M | A | A | M | L | A | D | E | R | L | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | T | V | R | K | V | A | D | A | C | I | A | S | I | A | Q | - | - | G | G | K | V | L | L | A | G | N | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | S | A | A | D | A | Q | H | I | A | G | Q | F | V | S | R | F | A | F | D | R | P | G | L | P | A | V | A | L | T | T | D | T | S | I | L | T | A | - | - | I | G | N | D | Y | G | Y | E | K | L | F | S | R | Q | V | Q | A | - | L | G | N | E | G | D | V | L | I | G | Y | S | T | S | G | K | S | P | N | I | L | A | A | F | R | E | A | K | A | K | G | M | T | C | V | G | F | T | G | N | R | G | G | E | M | R | E | - | - | - | - | - | - | - | - | - | L | C | - | - | - | D | L | L | L | E | V | P | S | - | - | A | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | P | K | I | Q | E | G | H | L | V | L | G | H | I | V | C | G | L | V | E | H | S | I | F | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1nria_ | A | P44862 | GO:0006040 |
| d1nria_ | A | P44862 | GO:0009254 |
| d1nria_ | A | P44862 | GO:0016803 |
| d1nria_ | A | P44862 | GO:0016829 |
| d1nria_ | A | P44862 | GO:0016835 |
| d1nria_ | A | P44862 | GO:0046348 |
| d1nria_ | A | P44862 | GO:0097173 |
| d1nria_ | A | P44862 | GO:0097175 |
| d1nria_ | A | P44862 | GO:0097367 |
| d1nria_ | A | P44862 | GO:1901135 |
| d1nria_ | A | P44862 | GO:0006040 |
| d1nria_ | A | P44862 | GO:0009254 |
| d1nria_ | A | P44862 | GO:0016803 |
| d1nria_ | A | P44862 | GO:0016829 |
| d1nria_ | A | P44862 | GO:0016835 |
| d1nria_ | A | P44862 | GO:0046348 |
| d1nria_ | A | P44862 | GO:0097173 |
| d1nria_ | A | P44862 | GO:0097175 |
| d1nria_ | A | P44862 | GO:0097367 |
| d1nria_ | A | P44862 | GO:1901135 |
| d1wiwa_ | A | Q5SIM3 | GO:0004347 |
| d1wiwa_ | A | Q5SIM3 | GO:0004476 |
| d1wiwa_ | A | Q5SIM3 | GO:0005975 |
| d1wiwa_ | A | Q5SIM3 | GO:0097367 |
| d1wiwa_ | A | Q5SIM3 | GO:1901135 |
| d1wiwa_ | A | Q5SIM3 | GO:0004347 |
| d1wiwa_ | A | Q5SIM3 | GO:0004476 |
| d1wiwa_ | A | Q5SIM3 | GO:0005975 |
| d1wiwa_ | A | Q5SIM3 | GO:0097367 |
| d1wiwa_ | A | Q5SIM3 | GO:1901135 |
