solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1di6a- ( 2 ) atLrIGLVSIsdd--------------------kGipALeeWLt
d1mkza1 ( 3 ) qvstefipTrIAILtVsnr------------gee-d--dtsGhyLrdsAq
d1uuya- ( 3 ) gpeykVAILtVsdt--vsag--------agp--drsgprAvsvVd
d1uz5a3 ( 181 ) pkVAVISTgneivppgnelkp-----gqiyDinGraLCdaIn
d1wu2a3 ( 181 ) vkpkVGIIItGselie---epseegfkegkivetnsi-LqglVe
d1y5ea1 ( 12 ) keVrCKIVTIsdt--rte-----------t--DkSGqllhelLk
d2g2ca1 ( 2 ) hiksAIIVVsdr--istg--------tre--nkAlplLqrls-
d2pjka- ( 13 ) pksLnFyVITIsts--ryek---llkkepiv--desGdiIkqlLi
d3pzya- ( 4 ) rsArVIIAsta--ss----------------dcGpiItewLa
d3tcra- ( 21 ) pvgrSlVVIVndr--tahg---------dq--dtsGplVtelLa
d6fgca2 ( 499 ) pvvAvMStgnellnpeddllp-----gkirDsnrstllatiq
bbbbbbb aaaaaaaaaa
60 70 80 90 100
d1di6a- ( 34 ) sALtt--pfel-etrlipDeqaiIeqtLceLVdemsChLVLTTgGtgpa-
d1mkza1 ( 39 ) ea-----gHhVvdkaivkenryaIraqVsaWiasddVqVVLITgGTglte
d1uuya- ( 36 ) sssekLggAkVvatavVpdeverIkdiLqkWSdvdeMdLILTLgGTgftp
d1uz5a3 ( 218 ) el-----gGeGifmgvArddkesLkalIekAVn--vGdvVVISgga----
d1wu2a3 ( 222 ) kf-----fGepilygvlpddesiik-etleKak--nEcdIvlIt------
d1y5ea1 ( 42 ) ea-----ghkvtsyeivkddkesIqqAVlaGyhkedVdVVlTNggtgitk
d2g2ca1 ( 33 ) -----dysyelisevvvpegydtVveaIatAlk-qgArFIITAGGTgira
d2pjka- ( 51 ) en-----ghkiigysLvpddkikIlkAFtdAlsideVdVIISTgGTgysp
d3pzya- ( 34 ) qq-----gFss-apevvadg-spVGeALr-Aidd-dVdVILTSgGTgiap
d3tcra- ( 52 ) ea-----gFvvdgvvvvendlseIqnaVntAvi-ggVdLVVTVgGTgvtp
d6fgca2 ( 536 ) eh-----gyptinlgivgdnpddllnalneGis--rAdviitSggvsmge
a bbbbbbbb aaaaaaaaaaaaa bbbbb
110 120 130 140 150
d1di6a- ( 81 ) rdvTPdATlaVAdrem---pgfgeqMrqIslhf---vptailsrQvGVir
d1mkza1 ( 84 ) gdqAPeALlplFdrev----gfgevFrl-sfee--igtstlqsrAvAGvA
d1uuya- ( 86 ) rdvTPeATkkVIeret---pgllfvMmqeslki---tpfamlarsaAGir
d1uz5a3 ( 262 ) -dltAsVIe-elg-eVkvhg------iaiq--------pGk--pTIiGvI
d1wu2a3 ( 266 ) -dyahkfV------nLlfhg------ttik--------pGr--pFGyGe-
d1y5ea1 ( 87 ) rdvTieAVsalLdkei---vgfgelFri-Syle-digssA-lsrAIGGti
d2g2ca1 ( 81 ) knqTPeATasfIhtrc---egleqqilihgg----------lsrGiVGVt
d2pjka- ( 96 ) tdiTVeTIrklFdrei---egfsdvFrlvsfndpevkaaayltkASAGii
d3pzya- ( 77 ) tdsTPdqTvavVdyli---pglaeaIr-sglp----vptsvlsrgvCGVA
d3tcra- ( 96 ) rdvAPeATqplLdrel---lgIaeaIrssGlaag--vteAGlSrGvAGiS
d6fgca2 ( 579 ) kdylkqvldidlhAqihfgr------vfmk--------pGl--pttfAtl
aaaaa bb aaaaa bbbb
160 170 180 190 200
d1di6a- ( 125 ) k----qALILNLpg-pksIkeTLeGvkdaegnvvvhGiFAsVP------y
d1mkza1 ( 129 ) n----kTLILA-pgstkacrTaWe-----n------iIapqLdArtrpcN
d1uuya- ( 130 ) g----sTLIINMpgnpnaVaeCMe------------aLlpaLk------h
d1uz5a3 ( 293 ) k----gkpVFgLPgyptsCltNFt-----l------lVvplLlralgreg
d1wu2a3 ( 292 ) -------kVfi-sgypvsvfaQFn-----l------fVkhaLAk-vgaqn
d1y5ea1 ( 133 ) g----rkVVFS-pgssgavrla-n-----k------lilpeLg------h
d2g2ca1 ( 125 ) grddhAALIVNApsssggItdTwa------------vIspvIp------n
d2pjka- ( 143 ) g----kKIVYLLpgspdaVklALk-----e------lIlpeVg------h
d3pzya- ( 121 ) g-----tLIVNLpgspggVrdGLg------------vLagvLd------h
d3tcra- ( 141 ) g----sTLVVNIAgsraAVrdGMa------------tLtpmAi------q
d6fgca2 ( 613 ) didgvrkiiFAlPgnpvsAvvtCn-----l------fvvpalrkmqgild
bbbbbb aaaaaaaaa aaaaaa
210 220
d1di6a- ( 165 ) cIqllegpyVeTapevvaafRparr
d1mkza1 ( 164 ) fhphLk
d1uuya- ( 158 ) aLkqi
d1uz5a3 ( 328 ) -k
d1wu2a3 ( 324 ) y
d1y5ea1 ( 162 ) itfel
d2g2ca1 ( 157 ) ifegLda
d2pjka- ( 172 ) lVylVrs
d3pzya- ( 149 ) aldqlag
d3tcra- ( 169 ) iieqlss
d6fgca2 ( 652 ) pr
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| MogA | MogA-like | Escherichia coli [TaxId: 562] | 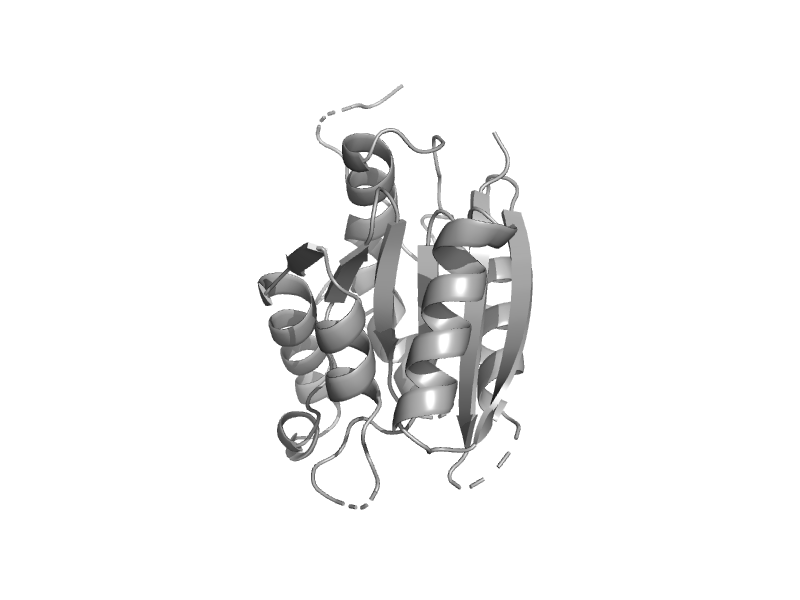 | view | |
| MoaB | MogA-like | Escherichia coli [TaxId: 562] | 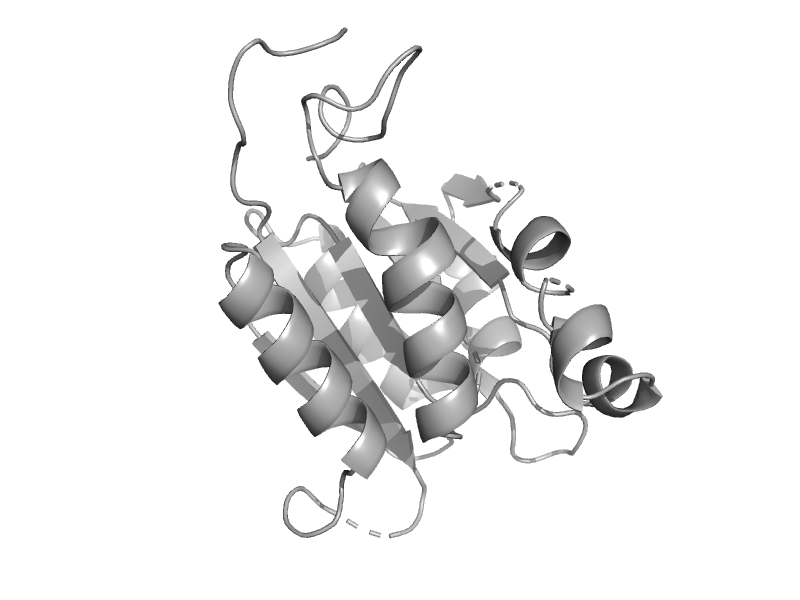 | view | |
| Plant CNX1 G domain | MogA-like | Thale cress (Arabidopsis thaliana) [TaxId: 3702] | 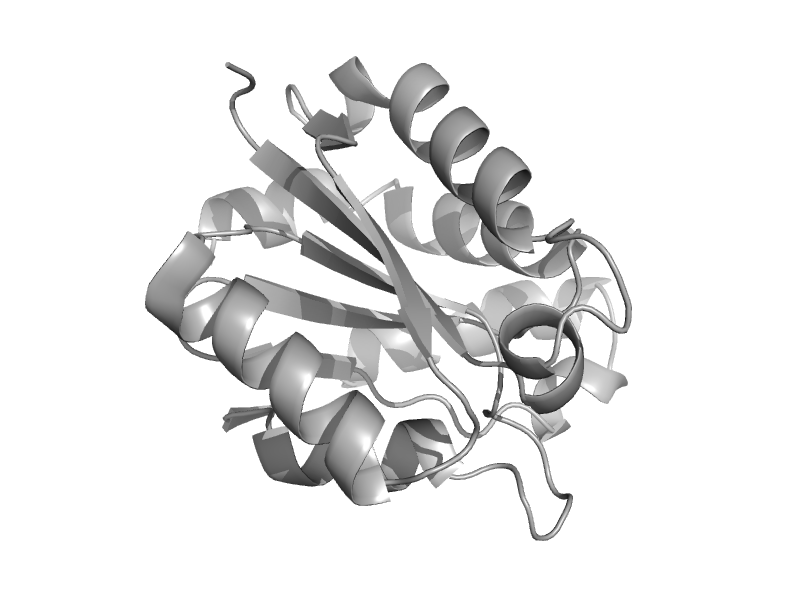 | view | |
| MoeA, central domain | MoeA central domain-like | Pyrococcus horikoshii, PH0582 [TaxId: 53953] | 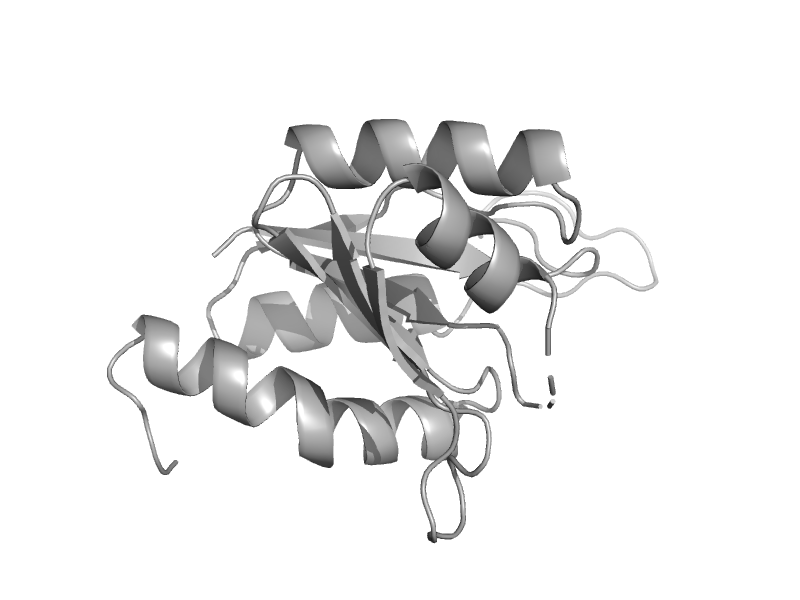 | view | |
| MoeA, central domain | MoeA central domain-like | Pyrococcus horikoshii, PH1647 [TaxId: 53953] |  | view | |
| MoaB | MogA-like | Bacillus cereus [TaxId: 1396] |  | view | |
| Putative molybdenum cofactor biosynthesis protein DIP0503 | MogA-like | Corynebacterium diphtheriae [TaxId: 1717] |  | view | |
| automated matches | automated matches | Sulfolobus tokodaii [TaxId: 111955] |  | view | |
| automated matches | automated matches | Mycobacterium avium [TaxId: 1770] |  | view | |
| automated matches | automated matches | Mycobacterium abscessus [TaxId: 561007] |  | view | |
| automated matches | automated matches | Norway rat (Rattus norvegicus) [TaxId: 10116] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | A | T | L | R | I | G | L | V | S | I | S | D | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | G | I | P | A | L | E | E | W | L | T | S | A | L | T | T | - | - | P | F | E | L | - | E | T | R | L | I | P | D | E | Q | A | I | I | E | Q | T | L | C | E | L | V | D | E | M | S | C | H | L | V | L | T | T | G | G | T | G | P | A | - | R | D | V | T | P | D | A | T | L | A | V | A | D | R | E | M | - | - | - | P | G | F | G | E | Q | M | R | Q | I | S | L | H | F | - | - | - | V | P | T | A | I | L | S | R | Q | V | G | V | I | R | K | - | - | - | - | Q | A | L | I | L | N | L | P | G | - | P | K | S | I | K | E | T | L | E | G | V | K | D | A | E | G | N | V | V | V | H | G | I | F | A | S | V | P | - | - | - | - | - | - | Y | C | I | Q | L | L | E | G | P | Y | V | E | T | A | P | E | V | V | A | A | F | R | P | A | R | R |
| Q | V | S | T | E | F | I | P | T | R | I | A | I | L | T | V | S | N | R | - | - | - | - | - | - | - | - | - | - | - | - | G | E | E | - | D | - | - | D | T | S | G | H | Y | L | R | D | S | A | Q | E | A | - | - | - | - | - | G | H | H | V | V | D | K | A | I | V | K | E | N | R | Y | A | I | R | A | Q | V | S | A | W | I | A | S | D | D | V | Q | V | V | L | I | T | G | G | T | G | L | T | E | G | D | Q | A | P | E | A | L | L | P | L | F | D | R | E | V | - | - | - | - | G | F | G | E | V | F | R | L | - | S | F | E | E | - | - | I | G | T | S | T | L | Q | S | R | A | V | A | G | V | A | N | - | - | - | - | K | T | L | I | L | A | - | P | G | S | T | K | A | C | R | T | A | W | E | - | - | - | - | - | N | - | - | - | - | - | - | I | I | A | P | Q | L | D | A | R | T | R | P | C | N | F | H | P | H | L | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | G | P | E | Y | K | V | A | I | L | T | V | S | D | T | - | - | V | S | A | G | - | - | - | - | - | - | - | - | A | G | P | - | - | D | R | S | G | P | R | A | V | S | V | V | D | S | S | S | E | K | L | G | G | A | K | V | V | A | T | A | V | V | P | D | E | V | E | R | I | K | D | I | L | Q | K | W | S | D | V | D | E | M | D | L | I | L | T | L | G | G | T | G | F | T | P | R | D | V | T | P | E | A | T | K | K | V | I | E | R | E | T | - | - | - | P | G | L | L | F | V | M | M | Q | E | S | L | K | I | - | - | - | T | P | F | A | M | L | A | R | S | A | A | G | I | R | G | - | - | - | - | S | T | L | I | I | N | M | P | G | N | P | N | A | V | A | E | C | M | E | - | - | - | - | - | - | - | - | - | - | - | - | A | L | L | P | A | L | K | - | - | - | - | - | - | H | A | L | K | Q | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | P | K | V | A | V | I | S | T | G | N | E | I | V | P | P | G | N | E | L | K | P | - | - | - | - | - | G | Q | I | Y | D | I | N | G | R | A | L | C | D | A | I | N | E | L | - | - | - | - | - | G | G | E | G | I | F | M | G | V | A | R | D | D | K | E | S | L | K | A | L | I | E | K | A | V | N | - | - | V | G | D | V | V | V | I | S | G | G | A | - | - | - | - | - | D | L | T | A | S | V | I | E | - | E | L | G | - | E | V | K | V | H | G | - | - | - | - | - | - | I | A | I | Q | - | - | - | - | - | - | - | - | P | G | K | - | - | P | T | I | I | G | V | I | K | - | - | - | - | G | K | P | V | F | G | L | P | G | Y | P | T | S | C | L | T | N | F | T | - | - | - | - | - | L | - | - | - | - | - | - | L | V | V | P | L | L | L | R | A | L | G | R | E | G | - | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | V | K | P | K | V | G | I | I | I | T | G | S | E | L | I | E | - | - | - | E | P | S | E | E | G | F | K | E | G | K | I | V | E | T | N | S | I | - | L | Q | G | L | V | E | K | F | - | - | - | - | - | F | G | E | P | I | L | Y | G | V | L | P | D | D | E | S | I | I | K | - | E | T | L | E | K | A | K | - | - | N | E | C | D | I | V | L | I | T | - | - | - | - | - | - | - | D | Y | A | H | K | F | V | - | - | - | - | - | - | N | L | L | F | H | G | - | - | - | - | - | - | T | T | I | K | - | - | - | - | - | - | - | - | P | G | R | - | - | P | F | G | Y | G | E | - | - | - | - | - | - | - | - | K | V | F | I | - | S | G | Y | P | V | S | V | F | A | Q | F | N | - | - | - | - | - | L | - | - | - | - | - | - | F | V | K | H | A | L | A | K | - | V | G | A | Q | N | Y | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | K | E | V | R | C | K | I | V | T | I | S | D | T | - | - | R | T | E | - | - | - | - | - | - | - | - | - | - | - | T | - | - | D | K | S | G | Q | L | L | H | E | L | L | K | E | A | - | - | - | - | - | G | H | K | V | T | S | Y | E | I | V | K | D | D | K | E | S | I | Q | Q | A | V | L | A | G | Y | H | K | E | D | V | D | V | V | L | T | N | G | G | T | G | I | T | K | R | D | V | T | I | E | A | V | S | A | L | L | D | K | E | I | - | - | - | V | G | F | G | E | L | F | R | I | - | S | Y | L | E | - | D | I | G | S | S | A | - | L | S | R | A | I | G | G | T | I | G | - | - | - | - | R | K | V | V | F | S | - | P | G | S | S | G | A | V | R | L | A | - | N | - | - | - | - | - | K | - | - | - | - | - | - | L | I | L | P | E | L | G | - | - | - | - | - | - | H | I | T | F | E | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | H | I | K | S | A | I | I | V | V | S | D | R | - | - | I | S | T | G | - | - | - | - | - | - | - | - | T | R | E | - | - | N | K | A | L | P | L | L | Q | R | L | S | - | - | - | - | - | - | D | Y | S | Y | E | L | I | S | E | V | V | V | P | E | G | Y | D | T | V | V | E | A | I | A | T | A | L | K | - | Q | G | A | R | F | I | I | T | A | G | G | T | G | I | R | A | K | N | Q | T | P | E | A | T | A | S | F | I | H | T | R | C | - | - | - | E | G | L | E | Q | Q | I | L | I | H | G | G | - | - | - | - | - | - | - | - | - | - | L | S | R | G | I | V | G | V | T | G | R | D | D | H | A | A | L | I | V | N | A | P | S | S | S | G | G | I | T | D | T | W | A | - | - | - | - | - | - | - | - | - | - | - | - | V | I | S | P | V | I | P | - | - | - | - | - | - | N | I | F | E | G | L | D | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | P | K | S | L | N | F | Y | V | I | T | I | S | T | S | - | - | R | Y | E | K | - | - | - | L | L | K | K | E | P | I | V | - | - | D | E | S | G | D | I | I | K | Q | L | L | I | E | N | - | - | - | - | - | G | H | K | I | I | G | Y | S | L | V | P | D | D | K | I | K | I | L | K | A | F | T | D | A | L | S | I | D | E | V | D | V | I | I | S | T | G | G | T | G | Y | S | P | T | D | I | T | V | E | T | I | R | K | L | F | D | R | E | I | - | - | - | E | G | F | S | D | V | F | R | L | V | S | F | N | D | P | E | V | K | A | A | A | Y | L | T | K | A | S | A | G | I | I | G | - | - | - | - | K | K | I | V | Y | L | L | P | G | S | P | D | A | V | K | L | A | L | K | - | - | - | - | - | E | - | - | - | - | - | - | L | I | L | P | E | V | G | - | - | - | - | - | - | H | L | V | Y | L | V | R | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | R | S | A | R | V | I | I | A | S | T | A | - | - | S | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | C | G | P | I | I | T | E | W | L | A | Q | Q | - | - | - | - | - | G | F | S | S | - | A | P | E | V | V | A | D | G | - | S | P | V | G | E | A | L | R | - | A | I | D | D | - | D | V | D | V | I | L | T | S | G | G | T | G | I | A | P | T | D | S | T | P | D | Q | T | V | A | V | V | D | Y | L | I | - | - | - | P | G | L | A | E | A | I | R | - | S | G | L | P | - | - | - | - | V | P | T | S | V | L | S | R | G | V | C | G | V | A | G | - | - | - | - | - | T | L | I | V | N | L | P | G | S | P | G | G | V | R | D | G | L | G | - | - | - | - | - | - | - | - | - | - | - | - | V | L | A | G | V | L | D | - | - | - | - | - | - | H | A | L | D | Q | L | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | P | V | G | R | S | L | V | V | I | V | N | D | R | - | - | T | A | H | G | - | - | - | - | - | - | - | - | - | D | Q | - | - | D | T | S | G | P | L | V | T | E | L | L | A | E | A | - | - | - | - | - | G | F | V | V | D | G | V | V | V | V | E | N | D | L | S | E | I | Q | N | A | V | N | T | A | V | I | - | G | G | V | D | L | V | V | T | V | G | G | T | G | V | T | P | R | D | V | A | P | E | A | T | Q | P | L | L | D | R | E | L | - | - | - | L | G | I | A | E | A | I | R | S | S | G | L | A | A | G | - | - | V | T | E | A | G | L | S | R | G | V | A | G | I | S | G | - | - | - | - | S | T | L | V | V | N | I | A | G | S | R | A | A | V | R | D | G | M | A | - | - | - | - | - | - | - | - | - | - | - | - | T | L | T | P | M | A | I | - | - | - | - | - | - | Q | I | I | E | Q | L | S | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | P | V | V | A | V | M | S | T | G | N | E | L | L | N | P | E | D | D | L | L | P | - | - | - | - | - | G | K | I | R | D | S | N | R | S | T | L | L | A | T | I | Q | E | H | - | - | - | - | - | G | Y | P | T | I | N | L | G | I | V | G | D | N | P | D | D | L | L | N | A | L | N | E | G | I | S | - | - | R | A | D | V | I | I | T | S | G | G | V | S | M | G | E | K | D | Y | L | K | Q | V | L | D | I | D | L | H | A | Q | I | H | F | G | R | - | - | - | - | - | - | V | F | M | K | - | - | - | - | - | - | - | - | P | G | L | - | - | P | T | T | F | A | T | L | D | I | D | G | V | R | K | I | I | F | A | L | P | G | N | P | V | S | A | V | V | T | C | N | - | - | - | - | - | L | - | - | - | - | - | - | F | V | V | P | A | L | R | K | M | Q | G | I | L | D | P | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
