solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1jf8a- ( 2 ) dkktIYFIStGNsarSQMAeGwGkeiLg-----egwnVySAGieth----
d1u2pa- ( 4 ) p-lhVTFVCtgNiCRSPMAekMFaqqLrhrglgdaVrVtSAGtgnw---h
d1y1la- ( 1 ) kVLFVCihNTARSVMAeaLFnamAk------swkAeSAGvekae---
d2gi4a1 ( 1 ) mkkilFiClgniCRSPMaefiMkdlvkkanlekeffinsAGtsGe---h
d2myna- ( 1 ) mkkvMFvCkrnSCrSqMAEGFAktlGa-----gkiavtSCGless----
d3jvia1 ( 1 ) mkLLFVClgNicrSPAAeAVMkkVIqnhhlte-yiCdSAGTcsh----
d3rh0a- ( 1 ) mksVLFV-vgNggkSQMAaaLAqkyAs-----dsveIhSAGtkpaq---
d3rofa1 ( 1 ) mvdvAFvClgniCRSPMAeAiMrqrlkdrni-hdikvhSrGtgSw---n
d4etna1 ( 1 ) mdiiFvCtgntsrSPMAealFksiAeregl--nvnVrSAGvfAs----
d5gota1 ( 1 ) mkkVCFVClgNicRSPMAEFVMksiVss----dvMmIeSrATsdw---e
d6u1pa- ( 5 ) glsvLVVCtgNlcRSPmaEIilrdkirq-kl--niqVrSAGTlktg---
d7dhfa1 ( 2 ) fnkILVVAvgNiCRSPTGerVLqklLp------nktVaSAGiaAeksrl
d7kh8a1 ( 4 ) atksVLFVClgNiCRSPIAEAVFrklVtdqnisenWrVdSAATsgy---e
bbbbbb aaaaaaaaaaaa bbbbbb
60 70 80 90 100
d1jf8a- ( 43 ) ---gvnpkAieAMkevdIdIs-nHtsdlidndiLkqSdLVVTLcsdAdnn
d1u2pa- ( 50 ) vgscAderAagvLrahgyptd--hrAaqVgtehla-AdLLVALdrnHarl
d1y1la- ( 39 ) ---rVdetVkrLLaerglkAk-e-kPrtvdeVnlddFdlIVTVÇee--ss
d2gi4a1 ( 47 ) dGeGmhyGtknKlaqlniehk-nftskkltqklCdeSdflitMDnsnfkn
d2myna- ( 41 ) ---rvhptAiamMeevgidis-gqtSdpienfnAddYdvviSlCgc----
d3jvia1 ( 47 ) -gqqAdsrMrkVGksrgyqVd--siSrpVvssDFknfdyIFAMdndNyye
d3rh0a- ( 42 ) ---glnqlSveSIaevgadMs-qgipkaidpellrtVdrVVILgddA---
d3rofa1 ( 46 ) lgepPhegtqkilnkhnipfd-gmiSelFeatdd--fdYivAMdqsnvdn
d4etna1 ( 43 ) pngkAtphAveAlfekhial--nhvSsplteelMesAdlvlAMthqHkqi
d5gota1 ( 43 ) hgnpIhsgTqsiLktyqInyditkcSkqItitdFntFdyIIGMdsdNvkn
d6u1pa- ( 49 ) -ktmPddkAlqaLqdygyhPm-vnpvqqVtqqdFieHdfIyAMdrtNlad
d7dhfa1 ( 45 ) igkpAdamAieVAkencVdVe-nHqSqqLtsalCsqydLILVMekghmea
d7kh8a1 ( 51 ) ignpPdyrGqsCMkrhgipMs--hvArqItkeDFatFdyILCMdesNlrd
aaaaaaaaa aaaa bbbb aaaa
110 120 130 140 150
d1jf8a- ( 89 ) Cpi-lpp----nVkkehw-------------gfddPagkewseFqrVrde
d1u2pa- ( 97 ) Lrql--gV-eaar-Vrm-LrsFdprsgthaldVedPyygdhsdFeeVFav
d1y1la- ( 82 ) çvv-Lp---Tdk-pvtrw-------------hIenPagkdegtYrrVLae
d2gi4a1 ( 96 ) vlknFt--ntqnk-vlk-itdfSp--slnydevPDpwysg--nfdetyki
d2myna- ( 83 ) gvn-lppeWvtqeifedW-------------qledPdgqslevFrtvrgq
d3jvia1 ( 94 ) LldrCpe-qykqk-Ifk-MvdFCtt---itteVpdPy--gekgFhrVIdI
d3rh0a- ( 85 ) qVd-mpesA--qgalerW-------------sIeepdaqgmerMriVrdq
d3rofa1 ( 93 ) iksinp---nlkgqlfk-llefSn---meesdvpdPyytn--nFegvydm
d4etna1 ( 91 ) iasqf-g-ryrdk-vft-lkeyvtgs---hgdvldPfggsidiYkqtRde
d5gota1 ( 93 ) Lkemsqh-qwdsk-Iyl-Fre---------ggVpdPwytn--dfeeTyql
d6u1pa- ( 97 ) LldiCpa-ehknk-Lal-flskanrq---ekevpdpyrrsseffqrtall
d7dhfa1 ( 94 ) LtqiA-p-eArgk-TmL-FGqwig-----qkdIpdPyrqskeaFvhAyql
d7kh8a1 ( 99 ) LnrksnqvktckAkIel-LGsYDpq---kqliIedPyygndsdFetVyqQ
bbb aaaaaaaa
160
d1jf8a- ( 121 ) IklaIekfklr
d1u2pa- ( 142 ) IesALpgLhdwVde-rlar
d1y1la- ( 114 ) IeerVkklvg--e
d2gi4a1 ( 138 ) lslACknllvflsk
d2myna- ( 119 ) vkervenliakis
d3jvia1 ( 139 ) LedACenLIikLeegkLin
d3rh0a- ( 119 ) IdnrVqalla
d3rofa1 ( 134 ) vlsSCdnlidyivkdanlk
d4etna1 ( 134 ) leellrqlAkqlkk
d5gota1 ( 129 ) VrkgCqdWlsrLmskey
d6u1pa- ( 141 ) iesgavalvdswq
d7dhfa1 ( 135 ) IdeAAqaWakkl
d7kh8a1 ( 145 ) CvrCCraFlekah
aaaaaaaaa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Arsenate reductase ArsC | Low-molecular-weight phosphotyrosine protein phosphatases | Staphylococcus aureus [TaxId: 1280] | 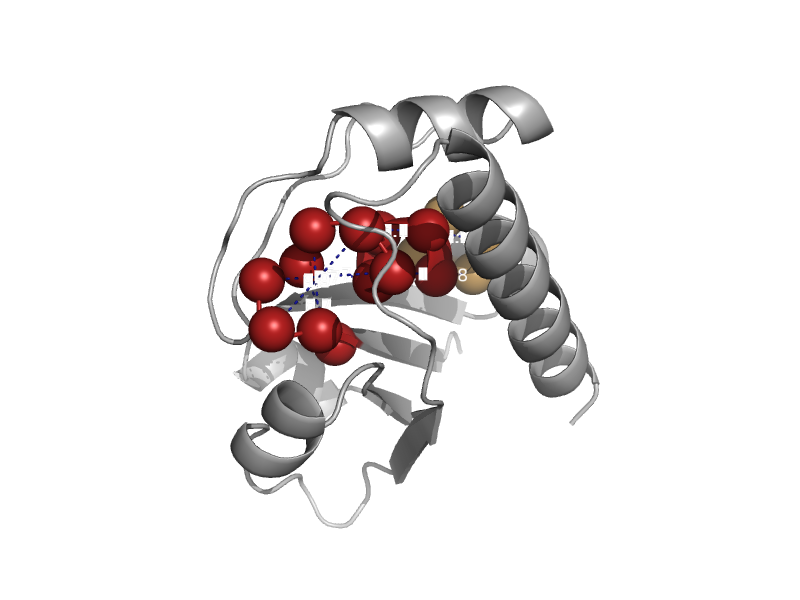 | view | |
| automated matches | automated matches | Mycobacterium tuberculosis [TaxId: 1773] | 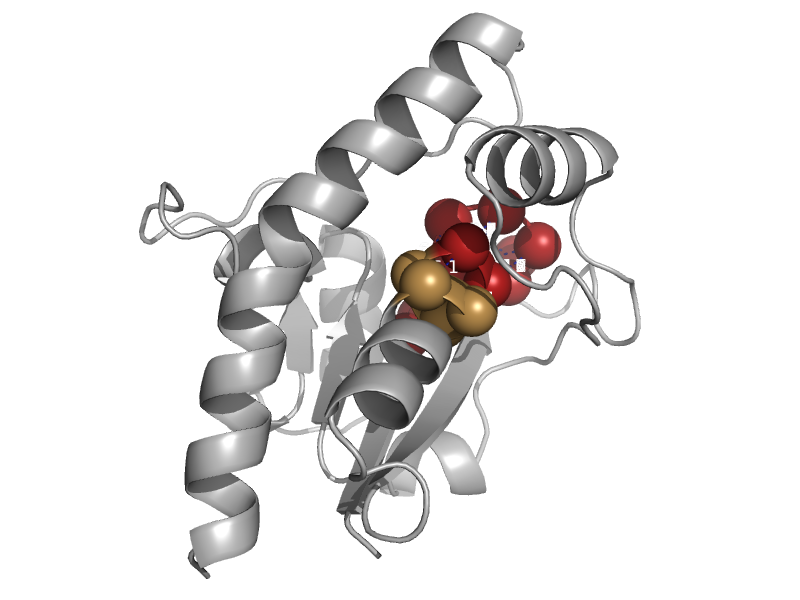 | view | |
| Arsenate reductase ArsC | Low-molecular-weight phosphotyrosine protein phosphatases | Archaeoglobus fulgidus [TaxId: 2234] | 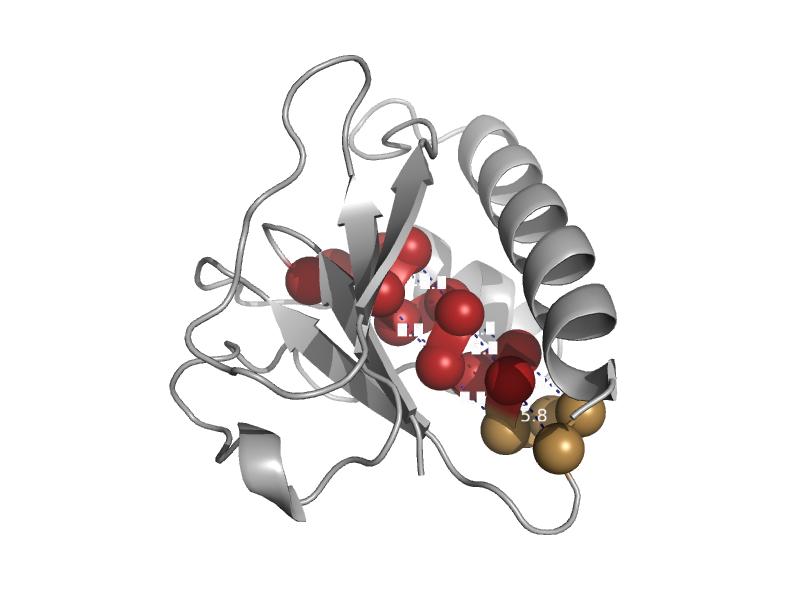 | view | |
| automated matches | automated matches | Campylobacter jejuni [TaxId: 197] | 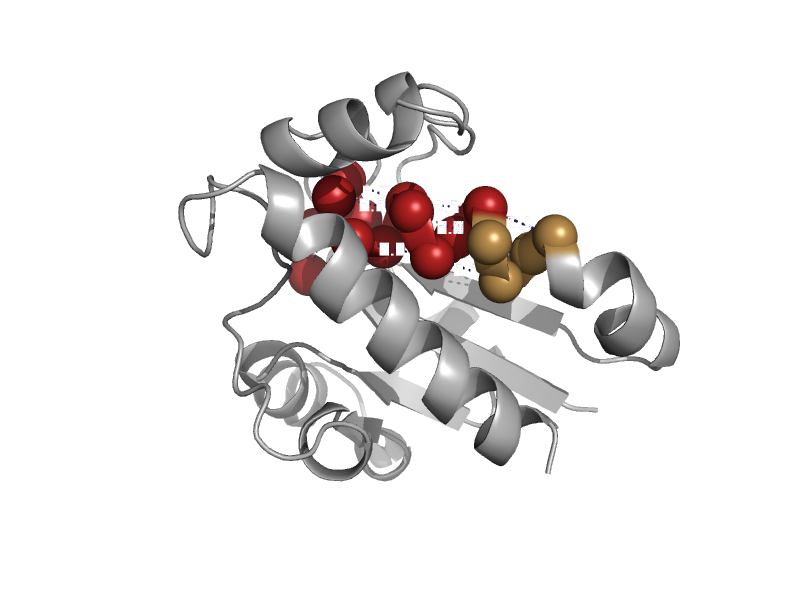 | view | |
| automated matches | automated matches | Synechocystis sp. [TaxId: 1111708] | 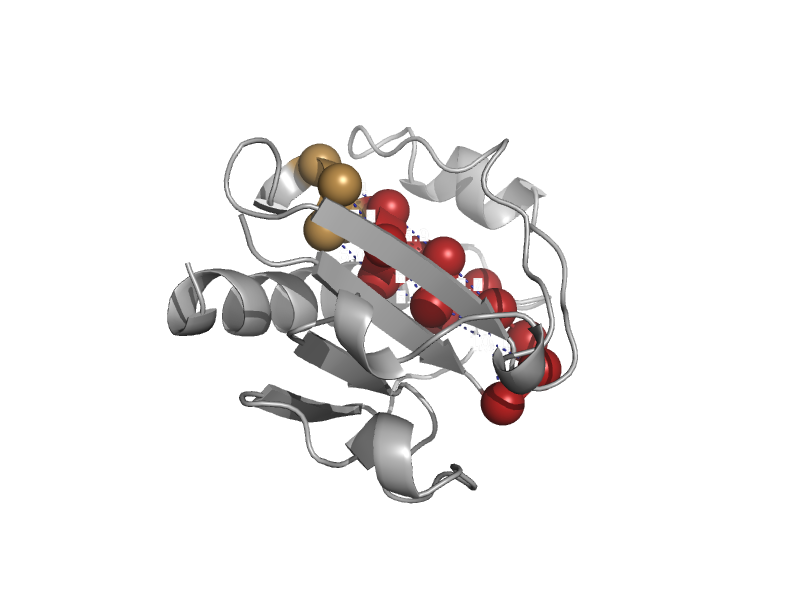 | view | |
| automated matches | automated matches | Entamoeba histolytica [TaxId: 294381] | 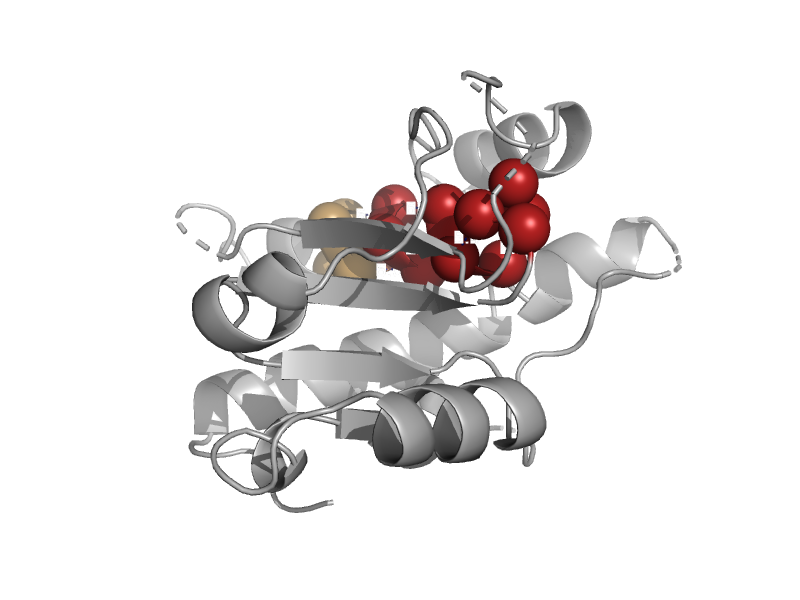 | view | |
| automated matches | automated matches | Corynebacterium glutamicum [TaxId: 1718] | 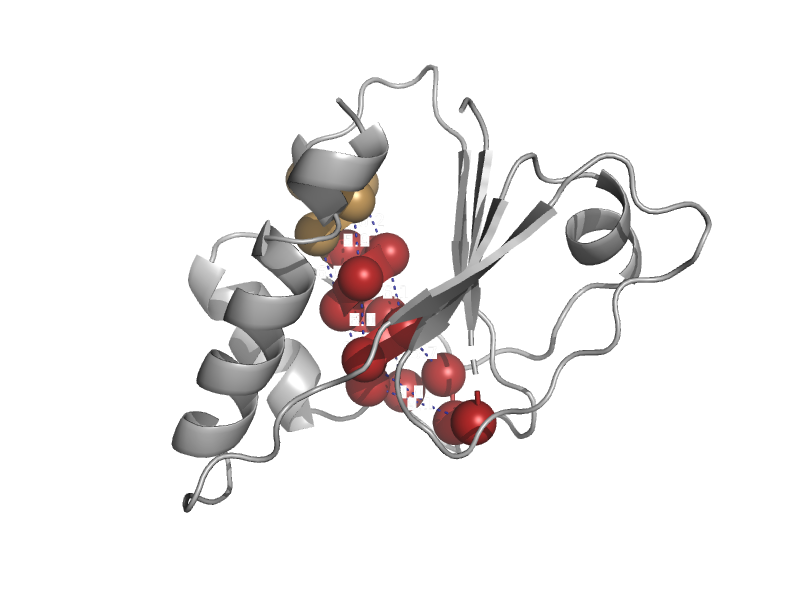 | view | |
| automated matches | automated matches | Staphylococcus aureus [TaxId: 1280] | 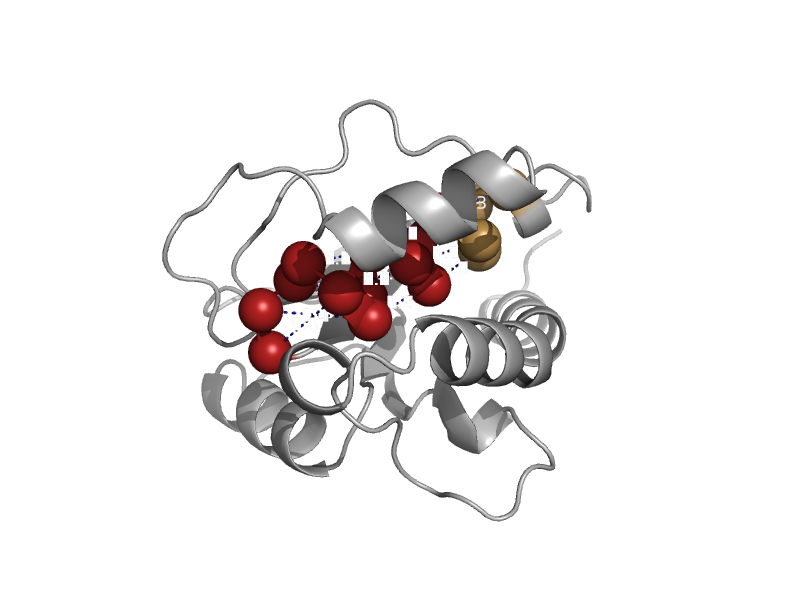 | view | |
| automated matches | automated matches | Bacillus subtilis [TaxId: 1423] |  | view | |
| automated matches | automated matches | Streptococcus pyogenes [TaxId: 1314] |  | view | |
| automated matches | automated matches | Vibrio cholerae [TaxId: 666] |  | view | |
| automated matches | automated matches | Vibrio vulnificus [TaxId: 672] |  | view | |
| Tyrosine phosphatase | Low-molecular-weight phosphotyrosine protein phosphatases | Human (Homo sapiens) [TaxId: 9606] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D | K | K | T | I | Y | F | I | S | T | G | N | S | A | R | S | Q | M | A | E | G | W | G | K | E | I | L | G | - | - | - | - | - | E | G | W | N | V | Y | S | A | G | I | E | T | H | - | - | - | - | - | - | - | G | V | N | P | K | A | I | E | A | M | K | E | V | D | I | D | I | S | - | N | H | T | S | D | L | I | D | N | D | I | L | K | Q | S | D | L | V | V | T | L | C | S | D | A | D | N | N | C | P | I | - | L | P | P | - | - | - | - | N | V | K | K | E | H | W | - | - | - | - | - | - | - | - | - | - | - | - | - | G | F | D | D | P | A | G | K | E | W | S | E | F | Q | R | V | R | D | E | I | K | L | A | I | E | K | F | K | L | R | - | - | - | - | - | - | - | - |
| P | - | L | H | V | T | F | V | C | T | G | N | I | C | R | S | P | M | A | E | K | M | F | A | Q | Q | L | R | H | R | G | L | G | D | A | V | R | V | T | S | A | G | T | G | N | W | - | - | - | H | V | G | S | C | A | D | E | R | A | A | G | V | L | R | A | H | G | Y | P | T | D | - | - | H | R | A | A | Q | V | G | T | E | H | L | A | - | A | D | L | L | V | A | L | D | R | N | H | A | R | L | L | R | Q | L | - | - | G | V | - | E | A | A | R | - | V | R | M | - | L | R | S | F | D | P | R | S | G | T | H | A | L | D | V | E | D | P | Y | Y | G | D | H | S | D | F | E | E | V | F | A | V | I | E | S | A | L | P | G | L | H | D | W | V | D | E | - | R | L | A | R |
| - | - | - | K | V | L | F | V | C | I | H | N | T | A | R | S | V | M | A | E | A | L | F | N | A | M | A | K | - | - | - | - | - | - | S | W | K | A | E | S | A | G | V | E | K | A | E | - | - | - | - | - | - | R | V | D | E | T | V | K | R | L | L | A | E | R | G | L | K | A | K | - | E | - | K | P | R | T | V | D | E | V | N | L | D | D | F | D | L | I | V | T | V | C | E | E | - | - | S | S | C | V | V | - | L | P | - | - | - | T | D | K | - | P | V | T | R | W | - | - | - | - | - | - | - | - | - | - | - | - | - | H | I | E | N | P | A | G | K | D | E | G | T | Y | R | R | V | L | A | E | I | E | E | R | V | K | K | L | V | G | - | - | E | - | - | - | - | - | - |
| - | M | K | K | I | L | F | I | C | L | G | N | I | C | R | S | P | M | A | E | F | I | M | K | D | L | V | K | K | A | N | L | E | K | E | F | F | I | N | S | A | G | T | S | G | E | - | - | - | H | D | G | E | G | M | H | Y | G | T | K | N | K | L | A | Q | L | N | I | E | H | K | - | N | F | T | S | K | K | L | T | Q | K | L | C | D | E | S | D | F | L | I | T | M | D | N | S | N | F | K | N | V | L | K | N | F | T | - | - | N | T | Q | N | K | - | V | L | K | - | I | T | D | F | S | P | - | - | S | L | N | Y | D | E | V | P | D | P | W | Y | S | G | - | - | N | F | D | E | T | Y | K | I | L | S | L | A | C | K | N | L | L | V | F | L | S | K | - | - | - | - | - |
| - | M | K | K | V | M | F | V | C | K | R | N | S | C | R | S | Q | M | A | E | G | F | A | K | T | L | G | A | - | - | - | - | - | G | K | I | A | V | T | S | C | G | L | E | S | S | - | - | - | - | - | - | - | R | V | H | P | T | A | I | A | M | M | E | E | V | G | I | D | I | S | - | G | Q | T | S | D | P | I | E | N | F | N | A | D | D | Y | D | V | V | I | S | L | C | G | C | - | - | - | - | G | V | N | - | L | P | P | E | W | V | T | Q | E | I | F | E | D | W | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | L | E | D | P | D | G | Q | S | L | E | V | F | R | T | V | R | G | Q | V | K | E | R | V | E | N | L | I | A | K | I | S | - | - | - | - | - | - |
| - | - | M | K | L | L | F | V | C | L | G | N | I | C | R | S | P | A | A | E | A | V | M | K | K | V | I | Q | N | H | H | L | T | E | - | Y | I | C | D | S | A | G | T | C | S | H | - | - | - | - | - | G | Q | Q | A | D | S | R | M | R | K | V | G | K | S | R | G | Y | Q | V | D | - | - | S | I | S | R | P | V | V | S | S | D | F | K | N | F | D | Y | I | F | A | M | D | N | D | N | Y | Y | E | L | L | D | R | C | P | E | - | Q | Y | K | Q | K | - | I | F | K | - | M | V | D | F | C | T | T | - | - | - | I | T | T | E | V | P | D | P | Y | - | - | G | E | K | G | F | H | R | V | I | D | I | L | E | D | A | C | E | N | L | I | I | K | L | E | E | G | K | L | I | N |
| - | M | K | S | V | L | F | V | - | V | G | N | G | G | K | S | Q | M | A | A | A | L | A | Q | K | Y | A | S | - | - | - | - | - | D | S | V | E | I | H | S | A | G | T | K | P | A | Q | - | - | - | - | - | - | G | L | N | Q | L | S | V | E | S | I | A | E | V | G | A | D | M | S | - | Q | G | I | P | K | A | I | D | P | E | L | L | R | T | V | D | R | V | V | I | L | G | D | D | A | - | - | - | Q | V | D | - | M | P | E | S | A | - | - | Q | G | A | L | E | R | W | - | - | - | - | - | - | - | - | - | - | - | - | - | S | I | E | E | P | D | A | Q | G | M | E | R | M | R | I | V | R | D | Q | I | D | N | R | V | Q | A | L | L | A | - | - | - | - | - | - | - | - | - |
| - | M | V | D | V | A | F | V | C | L | G | N | I | C | R | S | P | M | A | E | A | I | M | R | Q | R | L | K | D | R | N | I | - | H | D | I | K | V | H | S | R | G | T | G | S | W | - | - | - | N | L | G | E | P | P | H | E | G | T | Q | K | I | L | N | K | H | N | I | P | F | D | - | G | M | I | S | E | L | F | E | A | T | D | D | - | - | F | D | Y | I | V | A | M | D | Q | S | N | V | D | N | I | K | S | I | N | P | - | - | - | N | L | K | G | Q | L | F | K | - | L | L | E | F | S | N | - | - | - | M | E | E | S | D | V | P | D | P | Y | Y | T | N | - | - | N | F | E | G | V | Y | D | M | V | L | S | S | C | D | N | L | I | D | Y | I | V | K | D | A | N | L | K |
| - | - | M | D | I | I | F | V | C | T | G | N | T | S | R | S | P | M | A | E | A | L | F | K | S | I | A | E | R | E | G | L | - | - | N | V | N | V | R | S | A | G | V | F | A | S | - | - | - | - | P | N | G | K | A | T | P | H | A | V | E | A | L | F | E | K | H | I | A | L | - | - | N | H | V | S | S | P | L | T | E | E | L | M | E | S | A | D | L | V | L | A | M | T | H | Q | H | K | Q | I | I | A | S | Q | F | - | G | - | R | Y | R | D | K | - | V | F | T | - | L | K | E | Y | V | T | G | S | - | - | - | H | G | D | V | L | D | P | F | G | G | S | I | D | I | Y | K | Q | T | R | D | E | L | E | E | L | L | R | Q | L | A | K | Q | L | K | K | - | - | - | - | - |
| - | M | K | K | V | C | F | V | C | L | G | N | I | C | R | S | P | M | A | E | F | V | M | K | S | I | V | S | S | - | - | - | - | D | V | M | M | I | E | S | R | A | T | S | D | W | - | - | - | E | H | G | N | P | I | H | S | G | T | Q | S | I | L | K | T | Y | Q | I | N | Y | D | I | T | K | C | S | K | Q | I | T | I | T | D | F | N | T | F | D | Y | I | I | G | M | D | S | D | N | V | K | N | L | K | E | M | S | Q | H | - | Q | W | D | S | K | - | I | Y | L | - | F | R | E | - | - | - | - | - | - | - | - | - | G | G | V | P | D | P | W | Y | T | N | - | - | D | F | E | E | T | Y | Q | L | V | R | K | G | C | Q | D | W | L | S | R | L | M | S | K | E | Y | - | - |
| - | G | L | S | V | L | V | V | C | T | G | N | L | C | R | S | P | M | A | E | I | I | L | R | D | K | I | R | Q | - | K | L | - | - | N | I | Q | V | R | S | A | G | T | L | K | T | G | - | - | - | - | K | T | M | P | D | D | K | A | L | Q | A | L | Q | D | Y | G | Y | H | P | M | - | V | N | P | V | Q | Q | V | T | Q | Q | D | F | I | E | H | D | F | I | Y | A | M | D | R | T | N | L | A | D | L | L | D | I | C | P | A | - | E | H | K | N | K | - | L | A | L | - | F | L | S | K | A | N | R | Q | - | - | - | E | K | E | V | P | D | P | Y | R | R | S | S | E | F | F | Q | R | T | A | L | L | I | E | S | G | A | V | A | L | V | D | S | W | Q | - | - | - | - | - | - |
| - | F | N | K | I | L | V | V | A | V | G | N | I | C | R | S | P | T | G | E | R | V | L | Q | K | L | L | P | - | - | - | - | - | - | N | K | T | V | A | S | A | G | I | A | A | E | K | S | R | L | I | G | K | P | A | D | A | M | A | I | E | V | A | K | E | N | C | V | D | V | E | - | N | H | Q | S | Q | Q | L | T | S | A | L | C | S | Q | Y | D | L | I | L | V | M | E | K | G | H | M | E | A | L | T | Q | I | A | - | P | - | E | A | R | G | K | - | T | M | L | - | F | G | Q | W | I | G | - | - | - | - | - | Q | K | D | I | P | D | P | Y | R | Q | S | K | E | A | F | V | H | A | Y | Q | L | I | D | E | A | A | Q | A | W | A | K | K | L | - | - | - | - | - | - | - |
| A | T | K | S | V | L | F | V | C | L | G | N | I | C | R | S | P | I | A | E | A | V | F | R | K | L | V | T | D | Q | N | I | S | E | N | W | R | V | D | S | A | A | T | S | G | Y | - | - | - | E | I | G | N | P | P | D | Y | R | G | Q | S | C | M | K | R | H | G | I | P | M | S | - | - | H | V | A | R | Q | I | T | K | E | D | F | A | T | F | D | Y | I | L | C | M | D | E | S | N | L | R | D | L | N | R | K | S | N | Q | V | K | T | C | K | A | K | I | E | L | - | L | G | S | Y | D | P | Q | - | - | - | K | Q | L | I | I | E | D | P | Y | Y | G | N | D | S | D | F | E | T | V | Y | Q | Q | C | V | R | C | C | R | A | F | L | E | K | A | H | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1jf8a_ | A | P0A006 | GO:0004725 |
| d1jf8a_ | A | P0A006 | GO:0005737 |
| d1jf8a_ | A | P0A006 | GO:0016491 |
| d1jf8a_ | A | P0A006 | GO:0030612 |
| d1jf8a_ | A | P0A006 | GO:0046685 |
| d1jf8a_ | A | P0A006 | GO:0046872 |
| d1y1la_ | A | O28910 | GO:0046685 |
