solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1vema1 ( 418 ) tpvmQtIvVknVp--ttigDtVyITGnraeLg
d2j43a1 ( 5 ) ashhLrhfktlpageslgsLgLwVwgdVdqps
d2j43a2 ( 112 ) plkegyLrINYh-nqsghYdnlAVwTfkdVktpt
d2j73a- ( 5 ) ft-eTtIvVhYh-rydgkydgwNLwIwPvepvsq
d3bmva2 ( 579 ) ltgnqicVrFVVnnAs--TvygenVyLTGnvaeLg
d3njxa2 ( 251 ) yvaasgrgkVagtAsgAd---ssmdwvVhWynd----
d5ghla- ( 509 ) ç-ttptaVaVtFdLt-At--TtygEnIyLVGsisqLg
d6j33a1 ( 32 ) dvvvrlpdvavpgeavqAsarqAVIhLvdiag-dyatkNLylwnnetÇdA
bbbbbbb bbbbbb
60 70 80 90 100
d1vema1 ( 448 ) swdtkqyPi-qLyyds----hsnDwrgnvvLpa--ernIeFkAFIkskdg
d2j43a1 ( 38 ) k----dwpngAittkakkdd--ygyyldvpLaakhrqqVsYLINNk----
d2j43a2 ( 145 ) t----dwp-nGldLs-hkgh--ygAyvdVplk-egAneIGFLILdksktg
d2j73a- ( 37 ) ------eG-kayqFt-gedd--fgkvAvVkLp-mdLtkVGIiVrln----
d3bmva2 ( 612 ) nwdts-kAiGpMfnqvvyq--yptWyydVsVPa--gttIqFkFIkkngn-
d3njxa2 ( 281 ) ------aaqywtyTss------sGsFtSpaMkp---gtYtMvYyQg----
d5ghla- ( 542 ) dwets-dGi-aLsad-kytssdplwyvtvtLpA--gesFeYKFIriesdd
d6j33a1 ( 88 ) Lsapvadwndvsttptgsdk--yGPyWvIpL-tkesgçINViVRdgt---
bbbbbb bbbbbbbb
110 120 130 140
d1vema1 ( 491 ) --tvkswQt-iqqswnpVpl----kttshtssw
d2j43a1 ( 79 ) -ag--enlS-kdqhisll----t-pknevwIdeny-hahayr
d2j43a2 ( 186 ) dai---kvqpkdYlFkeL----d-nhtqVFVkdtdpkVynnpyyid
d2j73a- ( 72 ) ewq--akdvakdrfIeIk----d-gkaeVwIlqgveeIfyekp
d3bmva2 ( 656 ) --ti-twEggsnhtyt-Vpss---stgtvivnWqq
d3njxa2 ( 312 ) ---------eyaVatssvtVsagstttknIsgsvk
d5ghla- ( 587 ) --sv-ewEsdpnreyt-Vpqaçgtstatvtdtwr
d6j33a1 ( 132 ) -----nklIdsdLrVsFs----dftdrTVsViagnsavyd
bbbb bbbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| beta-amylase | Starch-binding domain | Bacillus cereus [TaxId: 1396] |  | view | |
| Pullulanase PulA | PUD-like | Streptococcus pyogenes [TaxId: 1314] |  | view | |
| Pullulanase PulA | PUD-like | Streptococcus pyogenes [TaxId: 1314] |  | view | |
| Pullulanase PulA | PUD-like | Thermotoga maritima [TaxId: 2336] |  | view | |
| Cyclodextrin glycosyltransferase, C-terminal domain | Starch-binding domain | Thermoanaerobacterium thermosulfurigenes, EM1 [TaxId: 33950] | 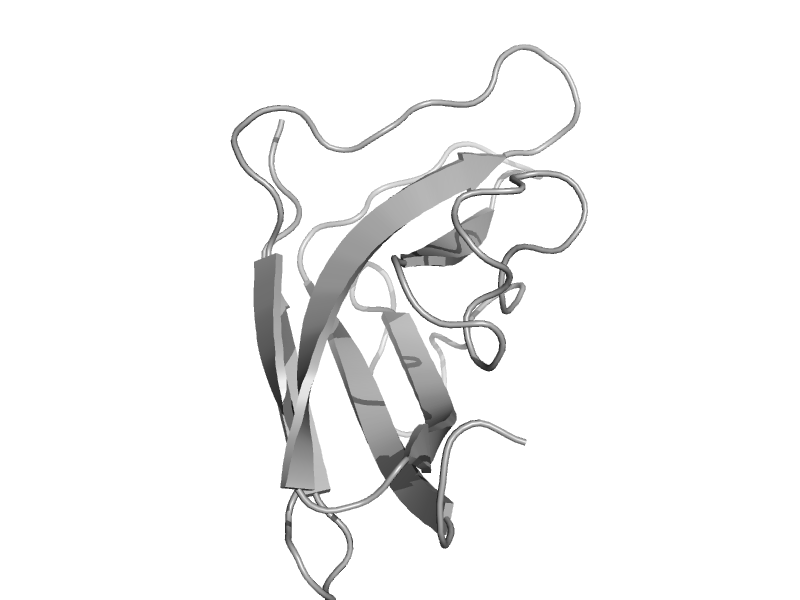 | view | |
| Rhamnogalacturonase B, RhgB, middle domain | Rhamnogalacturonase B, RhgB, middle domain | Fungus (Aspergillus aculeatus) [TaxId: 5053] | 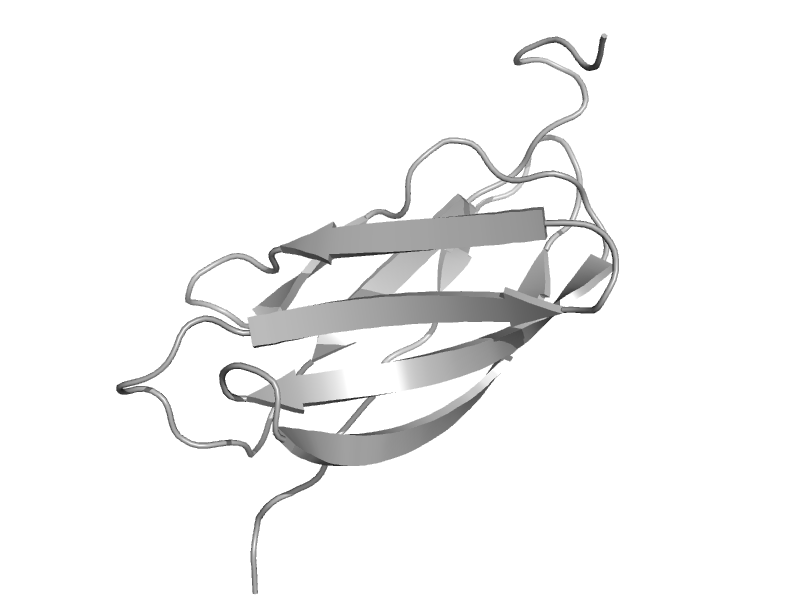 | view | |
| Glucoamylase, granular starch-binding domain | Starch-binding domain | Aspergillus niger [TaxId: 5061] | 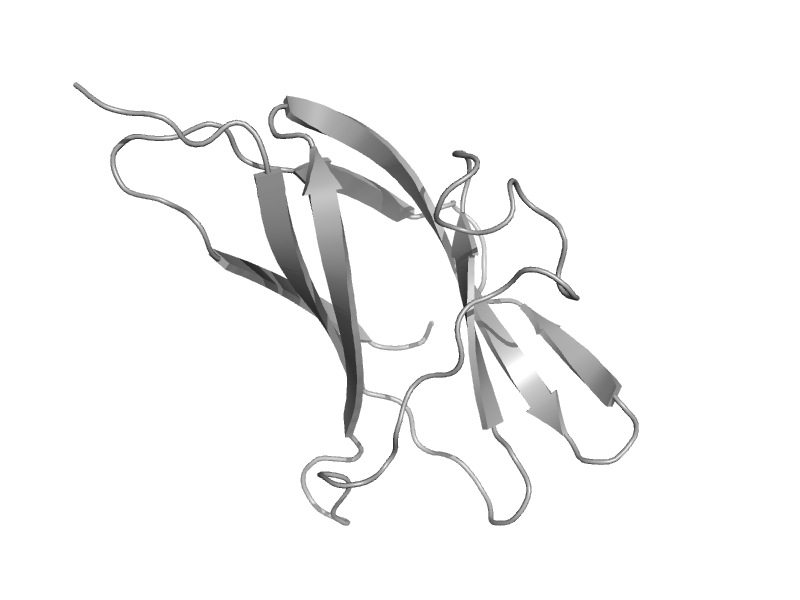 | view | |
| automated matches | automated matches | Klebsiella pneumoniae [TaxId: 573] | 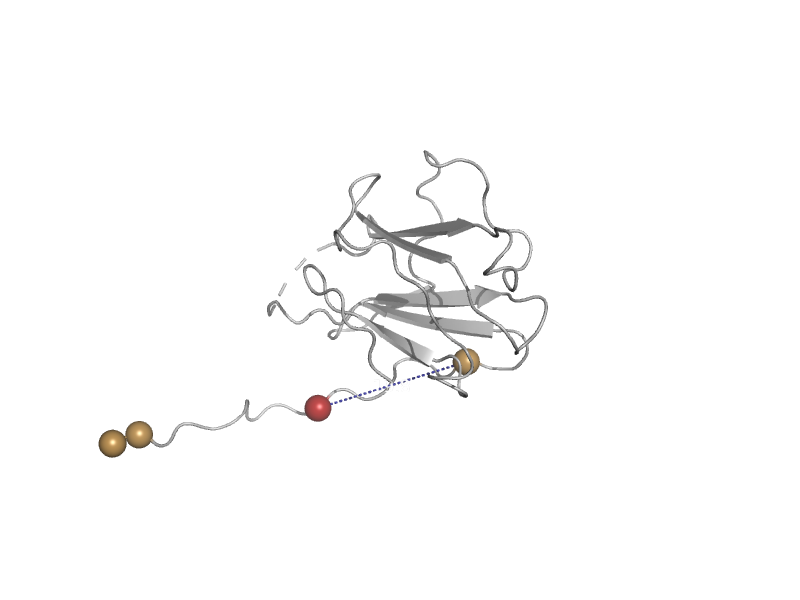 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | P | V | M | Q | T | I | V | V | K | N | V | P | - | - | T | T | I | G | D | T | V | Y | I | T | G | N | R | A | E | L | G | S | W | D | T | K | Q | Y | P | I | - | Q | L | Y | Y | D | S | - | - | - | - | H | S | N | D | W | R | G | N | V | V | L | P | A | - | - | E | R | N | I | E | F | K | A | F | I | K | S | K | D | G | - | - | T | V | K | S | W | Q | T | - | I | Q | Q | S | W | N | P | V | P | L | - | - | - | - | K | T | T | S | H | T | S | S | W | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | S | H | H | L | R | H | F | K | T | L | P | A | G | E | S | L | G | S | L | G | L | W | V | W | G | D | V | D | Q | P | S | K | - | - | - | - | D | W | P | N | G | A | I | T | T | K | A | K | K | D | D | - | - | Y | G | Y | Y | L | D | V | P | L | A | A | K | H | R | Q | Q | V | S | Y | L | I | N | N | K | - | - | - | - | - | A | G | - | - | E | N | L | S | - | K | D | Q | H | I | S | L | L | - | - | - | - | T | - | P | K | N | E | V | W | I | D | E | N | Y | - | H | A | H | A | Y | R | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | L | K | E | G | Y | L | R | I | N | Y | H | - | N | Q | S | G | H | Y | D | N | L | A | V | W | T | F | K | D | V | K | T | P | T | T | - | - | - | - | D | W | P | - | N | G | L | D | L | S | - | H | K | G | H | - | - | Y | G | A | Y | V | D | V | P | L | K | - | E | G | A | N | E | I | G | F | L | I | L | D | K | S | K | T | G | D | A | I | - | - | - | K | V | Q | P | K | D | Y | L | F | K | E | L | - | - | - | - | D | - | N | H | T | Q | V | F | V | K | D | T | D | P | K | V | Y | N | N | P | Y | Y | I | D |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | F | T | - | E | T | T | I | V | V | H | Y | H | - | R | Y | D | G | K | Y | D | G | W | N | L | W | I | W | P | V | E | P | V | S | Q | - | - | - | - | - | - | E | G | - | K | A | Y | Q | F | T | - | G | E | D | D | - | - | F | G | K | V | A | V | V | K | L | P | - | M | D | L | T | K | V | G | I | I | V | R | L | N | - | - | - | - | E | W | Q | - | - | A | K | D | V | A | K | D | R | F | I | E | I | K | - | - | - | - | D | - | G | K | A | E | V | W | I | L | Q | G | V | E | E | I | F | Y | E | K | P | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | T | G | N | Q | I | C | V | R | F | V | V | N | N | A | S | - | - | T | V | Y | G | E | N | V | Y | L | T | G | N | V | A | E | L | G | N | W | D | T | S | - | K | A | I | G | P | M | F | N | Q | V | V | Y | Q | - | - | Y | P | T | W | Y | Y | D | V | S | V | P | A | - | - | G | T | T | I | Q | F | K | F | I | K | K | N | G | N | - | - | - | T | I | - | T | W | E | G | G | S | N | H | T | Y | T | - | V | P | S | S | - | - | - | S | T | G | T | V | I | V | N | W | Q | Q | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | Y | V | A | A | S | G | R | G | K | V | A | G | T | A | S | G | A | D | - | - | - | S | S | M | D | W | V | V | H | W | Y | N | D | - | - | - | - | - | - | - | - | - | - | A | A | Q | Y | W | T | Y | T | S | S | - | - | - | - | - | - | S | G | S | F | T | S | P | A | M | K | P | - | - | - | G | T | Y | T | M | V | Y | Y | Q | G | - | - | - | - | - | - | - | - | - | - | - | - | - | E | Y | A | V | A | T | S | S | V | T | V | S | A | G | S | T | T | T | K | N | I | S | G | S | V | K | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | C | - | T | T | P | T | A | V | A | V | T | F | D | L | T | - | A | T | - | - | T | T | Y | G | E | N | I | Y | L | V | G | S | I | S | Q | L | G | D | W | E | T | S | - | D | G | I | - | A | L | S | A | D | - | K | Y | T | S | S | D | P | L | W | Y | V | T | V | T | L | P | A | - | - | G | E | S | F | E | Y | K | F | I | R | I | E | S | D | D | - | - | S | V | - | E | W | E | S | D | P | N | R | E | Y | T | - | V | P | Q | A | C | G | T | S | T | A | T | V | T | D | T | W | R | - | - | - | - | - | - | - | - | - | - | - | - |
| D | V | V | V | R | L | P | D | V | A | V | P | G | E | A | V | Q | A | S | A | R | Q | A | V | I | H | L | V | D | I | A | G | - | D | Y | A | T | K | N | L | Y | L | W | N | N | E | T | C | D | A | L | S | A | P | V | A | D | W | N | D | V | S | T | T | P | T | G | S | D | K | - | - | Y | G | P | Y | W | V | I | P | L | - | T | K | E | S | G | C | I | N | V | I | V | R | D | G | T | - | - | - | - | - | - | - | - | N | K | L | I | D | S | D | L | R | V | S | F | S | - | - | - | - | D | F | T | D | R | T | V | S | V | I | A | G | N | S | A | V | Y | D | - | - | - | - | - | - |
