solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d2j49a1 ( 149 ) penYirAYsmLknwVdssleiykpeLsyImYPIFIY
d2j4ba1 ( 18 ) qmetsYvsLktwIedsldlfkndLlpLLyPLFIH
d2nxpa1 ( 196 ) pdvsavlsaYnqqgdptyeeyYsgLkhfIecsldchraeLsqLfYPLFVH
aaaaaaaaaaaa aaaa aaaaaa
60 70 80 90 100
d2j49a1 ( 185 ) LFLnLVak-npvyArrFFdrFSpdfkdfhgseInrLfsVnsidhIkenev
d2j4ba1 ( 52 ) IYFdlIqqnktdeAkeFFekyrgdhk---seeIkqFesIytvqhIhennf
d2nxpa1 ( 248 ) -YlELVynqheneAksFfekFhgdQecyyqddLrvLssltkkeh-kgnet
aaaaa aaaaaaaaaaa333 aaaaa
110 120 130 140 150
d2j49a1 ( 234 ) AsafqshkyritMsktTlnlLlyfLnenesigGslIisVInqhLdpniv
d2j4ba1 ( 101 ) AytfknskyhlsMgryAfdlLinfLeern---ltyIlkILnqhLdikvyv
d2nxpa1 ( 298 ) ldfrTs-kfvLrIsrdSyqlLkrhLqekq---nNQiwnIVqehLyidifd
bbbb aaaaaaaaaaa aaaaaaaa bbbb
d2j49a1
d2j4ba1 ( 148 ) g
d2nxpa1
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| TAF5 subunit of TFIID | Taf5 N-terminal domain-like | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 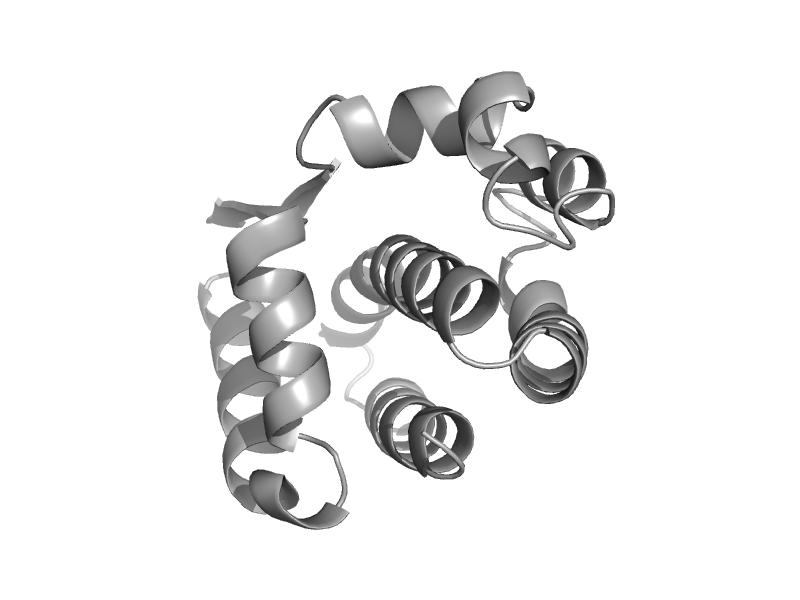 | view | |
| TAF5 subunit of TFIID | Taf5 N-terminal domain-like | Fungus (Encephalitozoon cuniculi) [TaxId: 6035] | 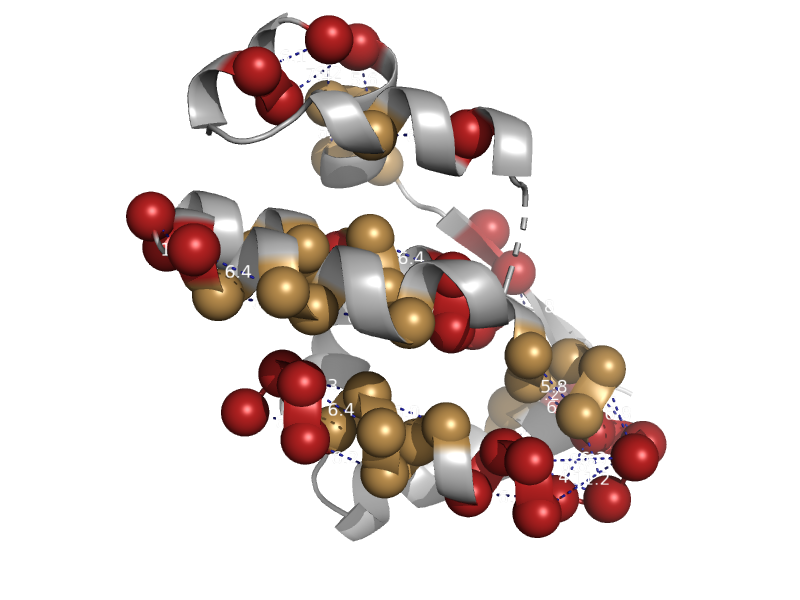 | view | |
| TAF5 subunit of TFIID | Taf5 N-terminal domain-like | Human (Homo sapiens) [TaxId: 9606] | 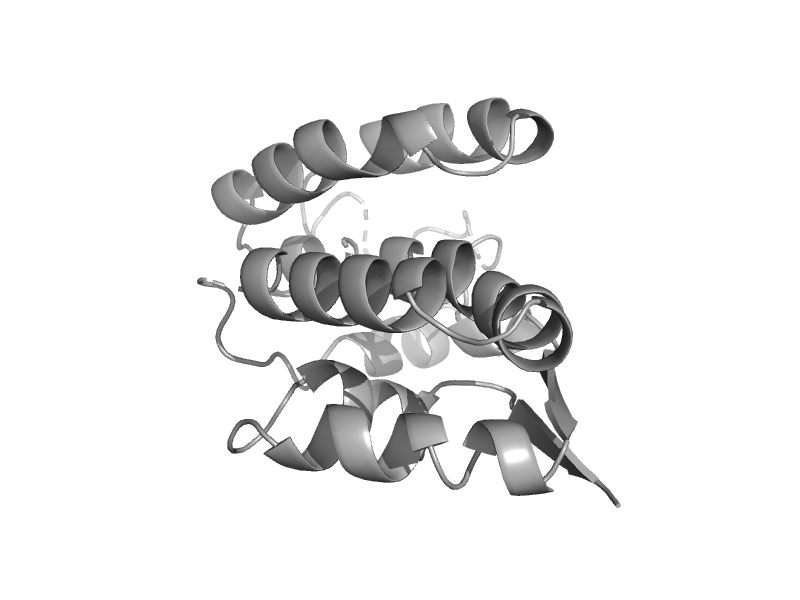 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | E | N | Y | I | R | A | Y | S | M | L | K | N | W | V | D | S | S | L | E | I | Y | K | P | E | L | S | Y | I | M | Y | P | I | F | I | Y | L | F | L | N | L | V | A | K | - | N | P | V | Y | A | R | R | F | F | D | R | F | S | P | D | F | K | D | F | H | G | S | E | I | N | R | L | F | S | V | N | S | I | D | H | I | K | E | N | E | V | A | S | A | F | Q | S | H | K | Y | R | I | T | M | S | K | T | T | L | N | L | L | L | Y | F | L | N | E | N | E | S | I | G | G | S | L | I | I | S | V | I | N | Q | H | L | D | P | N | I | V | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | M | E | T | S | Y | V | S | L | K | T | W | I | E | D | S | L | D | L | F | K | N | D | L | L | P | L | L | Y | P | L | F | I | H | I | Y | F | D | L | I | Q | Q | N | K | T | D | E | A | K | E | F | F | E | K | Y | R | G | D | H | K | - | - | - | S | E | E | I | K | Q | F | E | S | I | Y | T | V | Q | H | I | H | E | N | N | F | A | Y | T | F | K | N | S | K | Y | H | L | S | M | G | R | Y | A | F | D | L | L | I | N | F | L | E | E | R | N | - | - | - | L | T | Y | I | L | K | I | L | N | Q | H | L | D | I | K | V | Y | V | G |
| P | D | V | S | A | V | L | S | A | Y | N | Q | Q | G | D | P | T | Y | E | E | Y | Y | S | G | L | K | H | F | I | E | C | S | L | D | C | H | R | A | E | L | S | Q | L | F | Y | P | L | F | V | H | - | Y | L | E | L | V | Y | N | Q | H | E | N | E | A | K | S | F | F | E | K | F | H | G | D | Q | E | C | Y | Y | Q | D | D | L | R | V | L | S | S | L | T | K | K | E | H | - | K | G | N | E | T | L | D | F | R | T | S | - | K | F | V | L | R | I | S | R | D | S | Y | Q | L | L | K | R | H | L | Q | E | K | Q | - | - | - | N | N | Q | I | W | N | I | V | Q | E | H | L | Y | I | D | I | F | D | - |
| Member | Chain | UniProtKB | GO term |
|---|
