solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d2f66c1 ( 142 ) skkygdialkkkleqntkkldeeSsqletttr---si
d2f6ma2 ( 322 ) tdglnqlynlvaqdyaltdtiealsrmlhr-gti
d2f6mb2 ( 13 ) disqlfhdevplfdnsitskdkeVietlseIysivitLdhvekaylkdsi
d6vmee- ( 18 ) pelyeevklyk---na-erekydnmaeLfavVktMqalEkayikdcv
aaaaaaaaaaaaaaaaaaaaaaaaaaaa
60 70 80 90 100
d2f66c1 ( 176 ) dsaddldqfiknyldirtqyhlrreklatwd
d2f6ma2 ( 355 ) ----pldtfvkqgrelarqqflvrwhiq-ritspl---------------
d2f6mb2 ( 63 ) ----ddtqytntVdklLkqfkvylnsqnkeeInkhfqsieafadtynita
d6vmee- ( 62 ) ----spseYtaaCsrlLvqYkaafrqVqgseIssideFcrkf----rLdC
aaaaaaaaaaaaaaaaaaaaaa
110
d2f66c1
d2f6ma2 ( 385 ) -------------s
d2f6mb2 ( 109 ) snAitrlerg
d6vmee- ( 104 ) plAmerikedrpit
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Vacuolar protein sorting-associated protein 37, VPS37 (SRN2) | VPS37 C-terminal domain-like | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 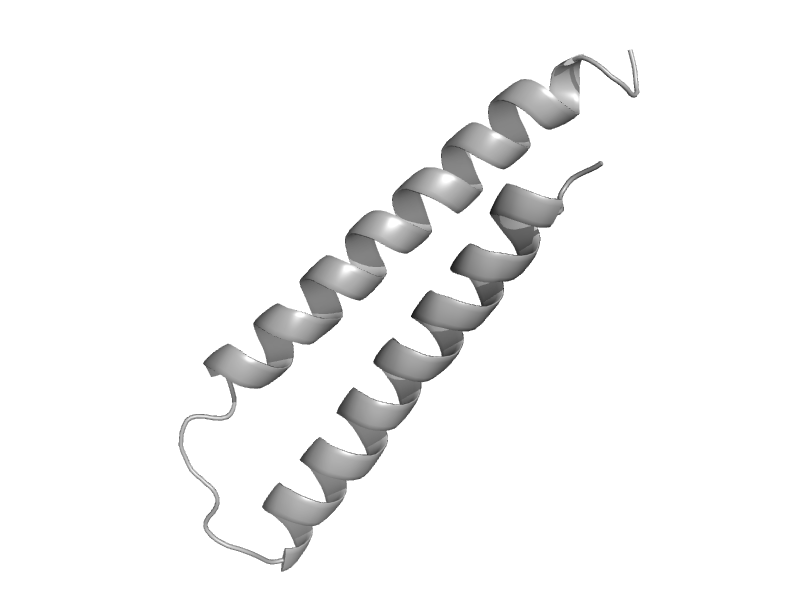 | view | |
| Vacuolar protein sorting-associated protein 23, VPS23 | VPS23 C-terminal domain | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 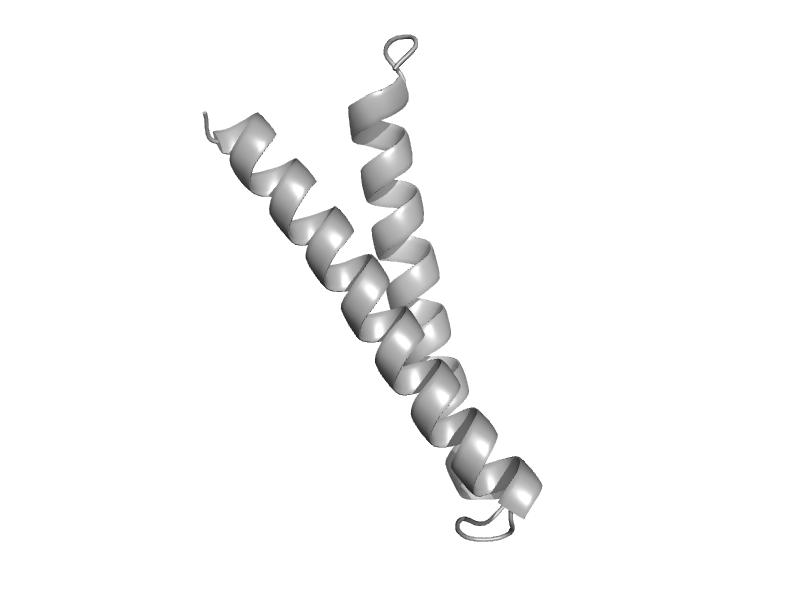 | view | |
| Vacuolar protein sorting-associated protein 28, VPS28 | VPS28 N-terminal domain | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 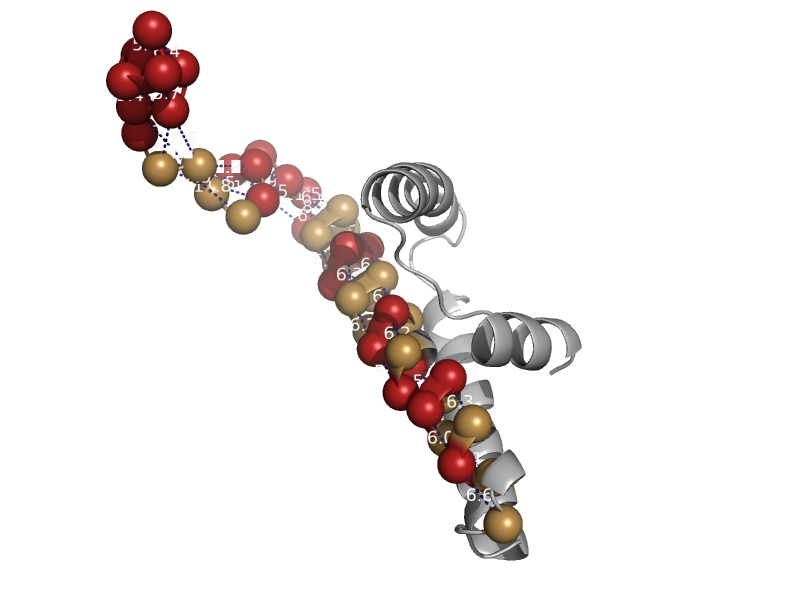 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 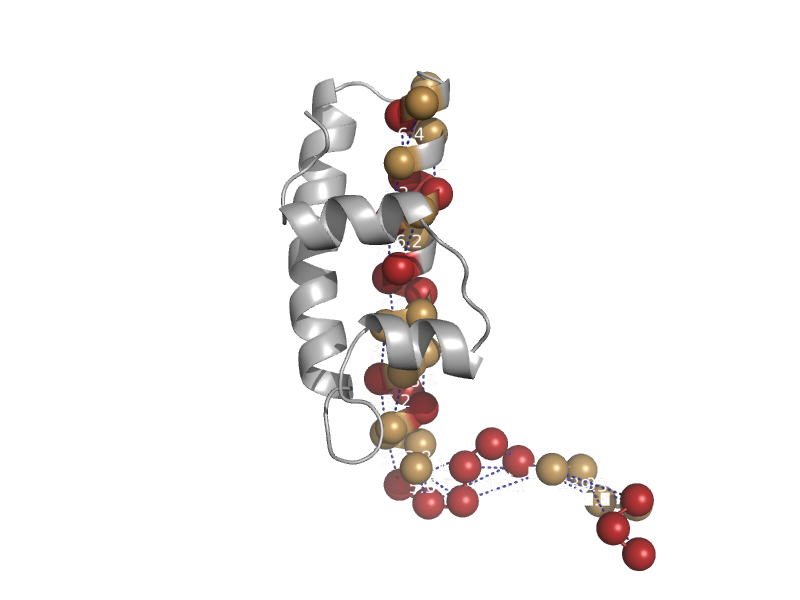 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | S | K | K | Y | G | D | I | A | L | K | K | K | L | E | Q | N | T | K | K | L | D | E | E | S | S | Q | L | E | T | T | T | R | - | - | - | S | I | D | S | A | D | D | L | D | Q | F | I | K | N | Y | L | D | I | R | T | Q | Y | H | L | R | R | E | K | L | A | T | W | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | D | G | L | N | Q | L | Y | N | L | V | A | Q | D | Y | A | L | T | D | T | I | E | A | L | S | R | M | L | H | R | - | G | T | I | - | - | - | - | P | L | D | T | F | V | K | Q | G | R | E | L | A | R | Q | Q | F | L | V | R | W | H | I | Q | - | R | I | T | S | P | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S |
| D | I | S | Q | L | F | H | D | E | V | P | L | F | D | N | S | I | T | S | K | D | K | E | V | I | E | T | L | S | E | I | Y | S | I | V | I | T | L | D | H | V | E | K | A | Y | L | K | D | S | I | - | - | - | - | D | D | T | Q | Y | T | N | T | V | D | K | L | L | K | Q | F | K | V | Y | L | N | S | Q | N | K | E | E | I | N | K | H | F | Q | S | I | E | A | F | A | D | T | Y | N | I | T | A | S | N | A | I | T | R | L | E | R | G | - | - | - | - |
| - | - | - | P | E | L | Y | E | E | V | K | L | Y | K | - | - | - | N | A | - | E | R | E | K | Y | D | N | M | A | E | L | F | A | V | V | K | T | M | Q | A | L | E | K | A | Y | I | K | D | C | V | - | - | - | - | S | P | S | E | Y | T | A | A | C | S | R | L | L | V | Q | Y | K | A | A | F | R | Q | V | Q | G | S | E | I | S | S | I | D | E | F | C | R | K | F | - | - | - | - | R | L | D | C | P | L | A | M | E | R | I | K | E | D | R | P | I | T |
| Member | Chain | UniProtKB | GO term |
|---|
