solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1ng6a- ( 1 ) msllerLnqdmklymknr-ekdkltVVrmVkasLqneaiklkkdsLtede
d1zq1c1 ( 457 ) elpqakVerYvkey---kldrslAqtlVd----ddlfeg-----------
d2d6fc1 ( 445 ) elpsekkerImrdy---gLsedlAsqLvkrn-lVdeFd-----------t
d2f2ab1 ( 294 ) elpderkakYvnel---gLpayDAhvLTltkemSdfFesTiehg---Adv
aaaaaaaaaa aaaaaaaaa
60 70 80 90 100
d1ng6a- ( 50 ) eltVLsreLkqrkdslqefsnanrldlvdkVqkeldiLevYLpeqlseee
d1zq1c1 ( 499 ) -paalv--------------------------------------------
d2d6fc1 ( 488 ) tviAsllAytlrelrr
d2f2ab1 ( 338 ) klTSnwLmggVneylnkn-------------qveLldtk------Ltpen
aaaaa aaaaa
110 120 130 140
d1ng6a- ( 100 ) LrtiVneTiaevgAsskadmgkVmgaImpkVkGkadgslInkLVssqLs
d1zq1c1 ( 512 ) ------------------------------------------------g
d2d6fc1
d2f2ab1 ( 369 ) LAgMIklied------gtMsskiAkkVFpeLAakggnAkqiMednglvq
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Hypothetical protein YqeY | GatB/YqeY domain | Bacillus subtilis [TaxId: 1423] | 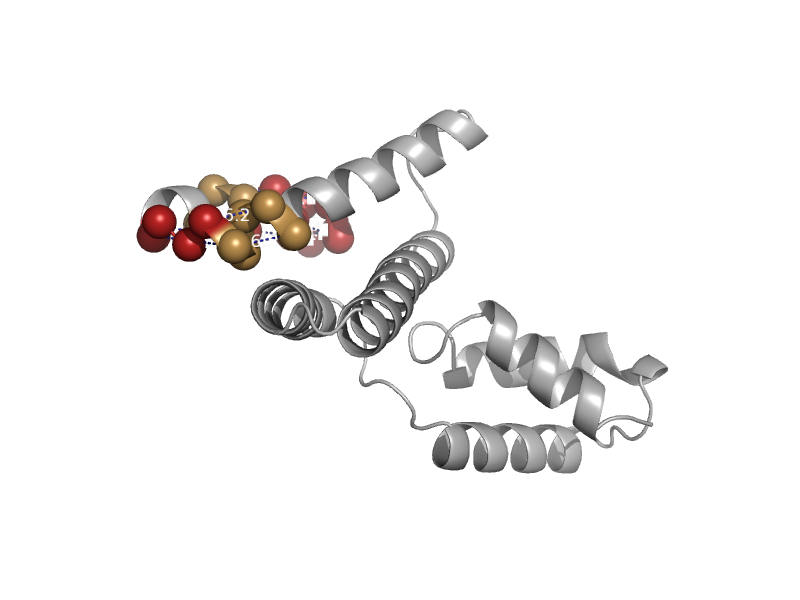 | view | |
| Glutamyl-tRNA(Gln) amidotransferase subunit E, GatE, C-terminal domain | GatB/GatE C-terminal domain-like | Pyrococcus abyssi [TaxId: 29292] |  | view | |
| Glutamyl-tRNA(Gln) amidotransferase subunit E, GatE, C-terminal domain | GatB/GatE C-terminal domain-like | Methanobacterium thermoautotrophicum [TaxId: 145262] |  | view | |
| Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, GatB, C-terminal domain | GatB/GatE C-terminal domain-like | Staphylococcus aureus [TaxId: 1280] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | S | L | L | E | R | L | N | Q | D | M | K | L | Y | M | K | N | R | - | E | K | D | K | L | T | V | V | R | M | V | K | A | S | L | Q | N | E | A | I | K | L | K | K | D | S | L | T | E | D | E | E | L | T | V | L | S | R | E | L | K | Q | R | K | D | S | L | Q | E | F | S | N | A | N | R | L | D | L | V | D | K | V | Q | K | E | L | D | I | L | E | V | Y | L | P | E | Q | L | S | E | E | E | L | R | T | I | V | N | E | T | I | A | E | V | G | A | S | S | K | A | D | M | G | K | V | M | G | A | I | M | P | K | V | K | G | K | A | D | G | S | L | I | N | K | L | V | S | S | Q | L | S |
| E | L | P | Q | A | K | V | E | R | Y | V | K | E | Y | - | - | - | K | L | D | R | S | L | A | Q | T | L | V | D | - | - | - | - | D | D | L | F | E | G | - | - | - | - | - | - | - | - | - | - | - | - | P | A | A | L | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G |
| E | L | P | S | E | K | K | E | R | I | M | R | D | Y | - | - | - | G | L | S | E | D | L | A | S | Q | L | V | K | R | N | - | L | V | D | E | F | D | - | - | - | - | - | - | - | - | - | - | - | T | T | V | I | A | S | L | L | A | Y | T | L | R | E | L | R | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| E | L | P | D | E | R | K | A | K | Y | V | N | E | L | - | - | - | G | L | P | A | Y | D | A | H | V | L | T | L | T | K | E | M | S | D | F | F | E | S | T | I | E | H | G | - | - | - | A | D | V | K | L | T | S | N | W | L | M | G | G | V | N | E | Y | L | N | K | N | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | V | E | L | L | D | T | K | - | - | - | - | - | - | L | T | P | E | N | L | A | G | M | I | K | L | I | E | D | - | - | - | - | - | - | G | T | M | S | S | K | I | A | K | K | V | F | P | E | L | A | A | K | G | G | N | A | K | Q | I | M | E | D | N | G | L | V | Q |
