solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d2qq4a- ( 1 ) msvldelyreilldHyqsprnfgvLp
d3a7ra2 ( 247 ) q
d3g0ma- ( 1 ) maaLpdkekLlrnFtrcanweekylyIieLGq--rLaeLnp
d4lw4c- ( 3 ) npqfagHpFgttv-taetLrntFapltqwedKyrqLimlGk--qLpaLpd
d5idha2 ( 248 ) k
d5ij6a2 ( 250 ) e
d5ufta1 ( 1 ) middiynrIl-fagnmerig-la
d6a6fa1 ( 1 ) miysefImdySklkkfhgkie
d7c8na- ( 1 ) miqqtg-yskkVmeHfmnprnvgvid
60 70 80 90 100
d2qq4a- ( 27 ) qatkqaggmnpscgDqVeVMVllegdtIadIrFqgqgcaistASASLMTe
d3a7ra2 ( 248 ) apafshlLderftwggVeLhFdVekghItrAqVftdSl--npaPLeaLag
d3g0ma- ( 40 ) qDrnp-ntIhg-CqsqVwIVMrrnangiIeLqGdSd-aaiVkGLMAVVFI
d4lw4c- ( 50 ) elkaqAkeiag-cenrVwLgytvaengkMHFFGdSe-grivrGLLAVLLT
d5idha2 ( 249 ) spafnlerrhrfpigsIeMkMnvadgaIqeIkIfgdff--glgeikdVed
d5ij6a2 ( 251 ) apkftikkeekfkggiVdArLTvekgkIieLtIygdyf--akketteIva
d5ufta1 ( 25 ) epdavAtvs--kcgstVtVyLkMrdgvVtdFaHeV-AcaLGqASSSVMAr
d6a6fa1 ( 22 ) nAhkveegknlscgDeVtLyFlfdgdkIvdVkFeghgcaISqASTNVMIe
d7c8na- ( 26 ) dpdGygkvgNpvcgdlMeIfIkVgdekIedIkFrTfGcgAAiATSSMITe
bbb bbbbbbbb bbbbbbbbb aaaaaaaa
110 120 130 140 150
d2qq4a- ( 77 ) aVkgkk--vaeAlelSrkFqaMVvegappdp-tLgdLlALqgVaklpa-r
d3a7ra2 ( 296 ) rLqgclyrAdmLqqeCe-----------------aLlvdfpeq-------
d3g0ma- ( 88 ) LYhqmt--aqdIvhfd----------------vrpwfekMaLaqhLtpsr
d4lw4c- ( 98 ) aVegkt--aaeLqaqs----------------plalFdeLgLraqLsaSr
d5idha2 ( 297 ) iLtgvkydkasLeeaId-----------------qIdVkkyFg-------
d5ij6a2 ( 299 ) aLlgvdyqyssIwqaLa-----------------afnFedYFv-------
d5ufta1 ( 75 ) nVigAt--adeLraardaMyrMLkengpaPegrFadMkyFepVrdy-a-r
d6a6fa1 ( 72 ) qIigkt--kqeAleMMkNAenMMl-gkefdenvLgpIinFydVknypm-r
d7c8na- ( 76 ) mArgks--LeeAmritrndva----------------daLdgLpp--q-k
aa aaaaaa
160 170
d2qq4a- ( 123 ) vkcATLAWhALeeALr
d3a7ra2 ( 322 ) ekeLreLSawMAgaVr
d3g0ma- ( 120 ) sqGLeaMIraIrakAatls
d4lw4c- ( 130 ) sqgLnaLseaIiaAAkqv
d5idha2 ( 323 ) nIekedLlgLIy
d5ij6a2 ( 325 ) nItkeeFVhlLvd
d5ufta1 ( 122 ) haSTlLTFdAVadCIrqie---a
d6a6fa1 ( 118 ) vkCFllPWkTLeiALk
d7c8na- ( 105 ) macSnlAAdALhaAIndylskkq
aaaaaaaaaaaaaa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| automated matches | automated matches | Thermus thermophilus [TaxId: 274] | 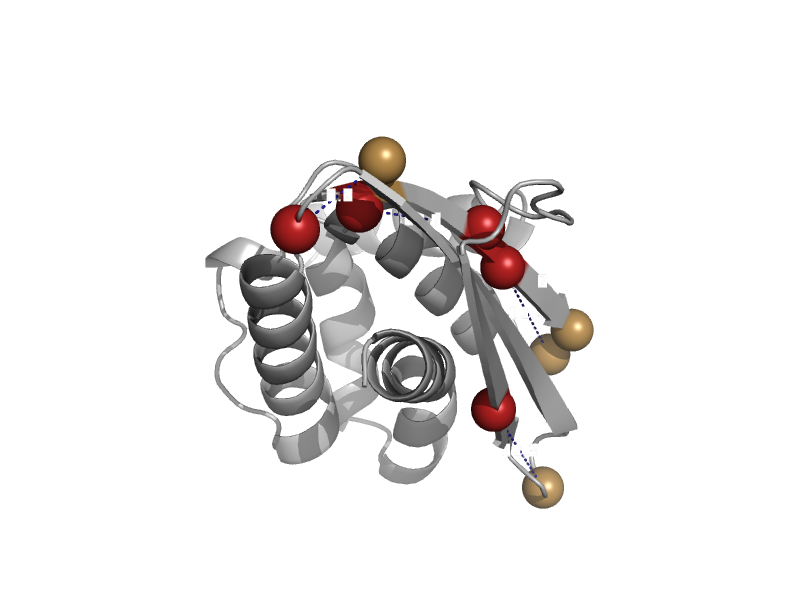 | view | |
| Two-domain LplA, C-terminal domain | SP1160 C-terminal domain-like | Escherichia coli [TaxId: 562] | 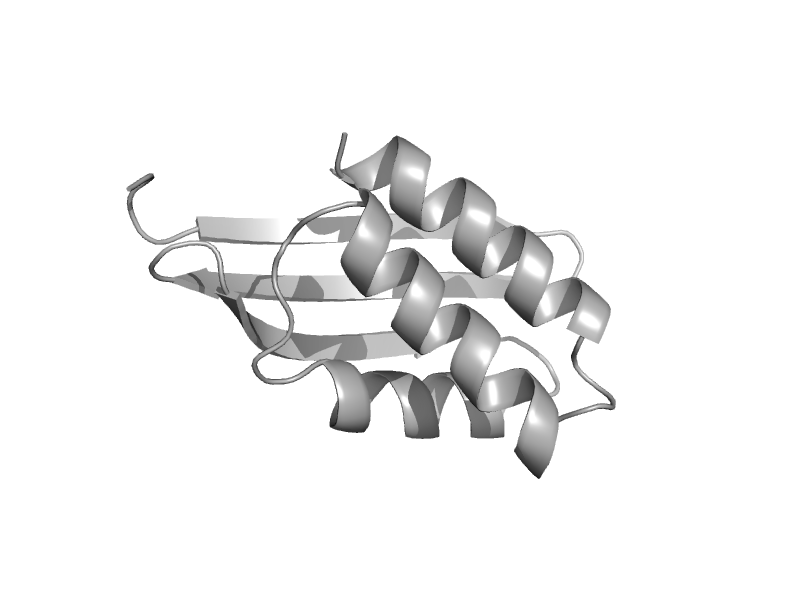 | view | |
| automated matches | SufE-like | Salmonella typhimurium [TaxId: 99287] | 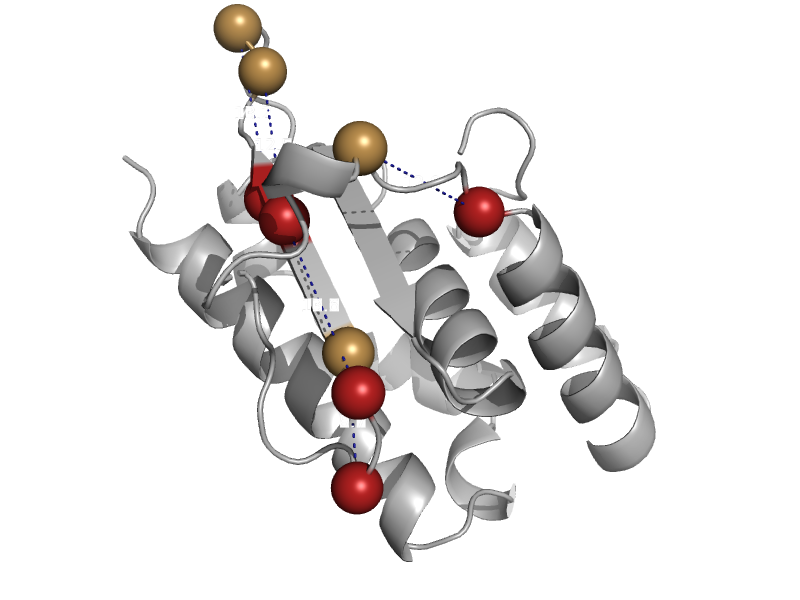 | view | |
| automated matches | SufE-like | Escherichia coli [TaxId: 714962] | 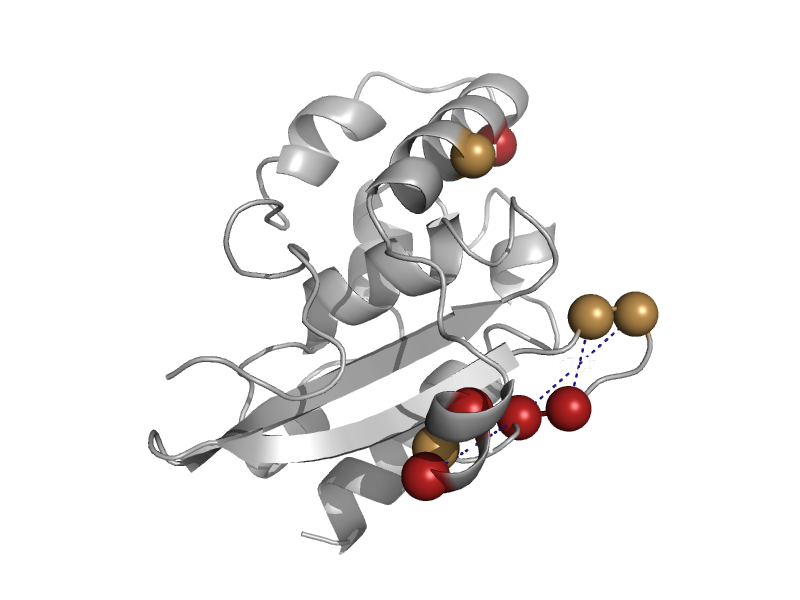 | view | |
| automated matches | automated matches | Enterococcus faecalis [TaxId: 226185] | 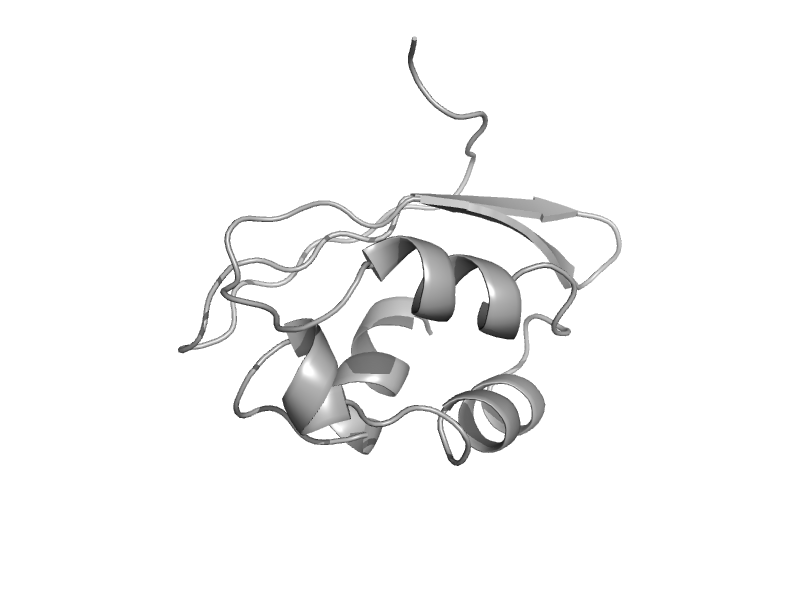 | view | |
| automated matches | automated matches | Enterococcus faecalis [TaxId: 226185] | 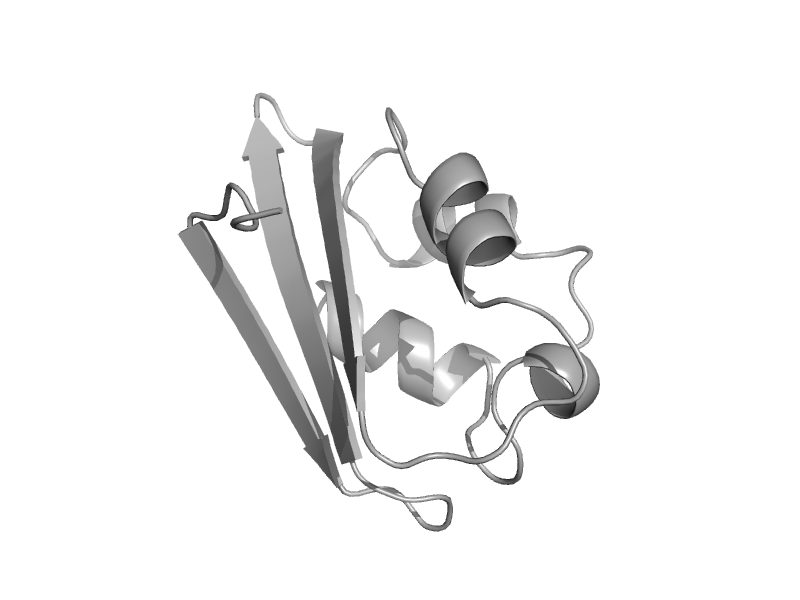 | view | |
| automated matches | automated matches | Brucella abortus [TaxId: 359391] | 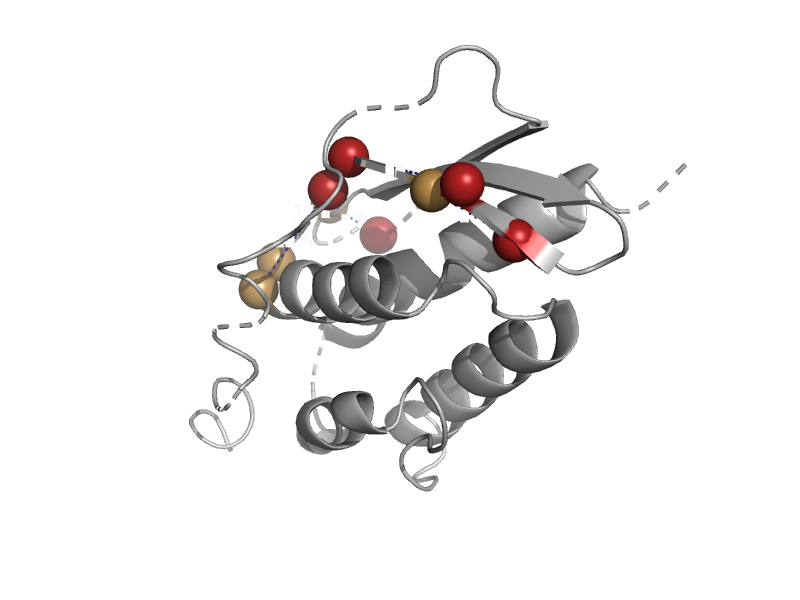 | view | |
| automated matches | automated matches | Fervidobacterium islandicum [TaxId: 2423] | 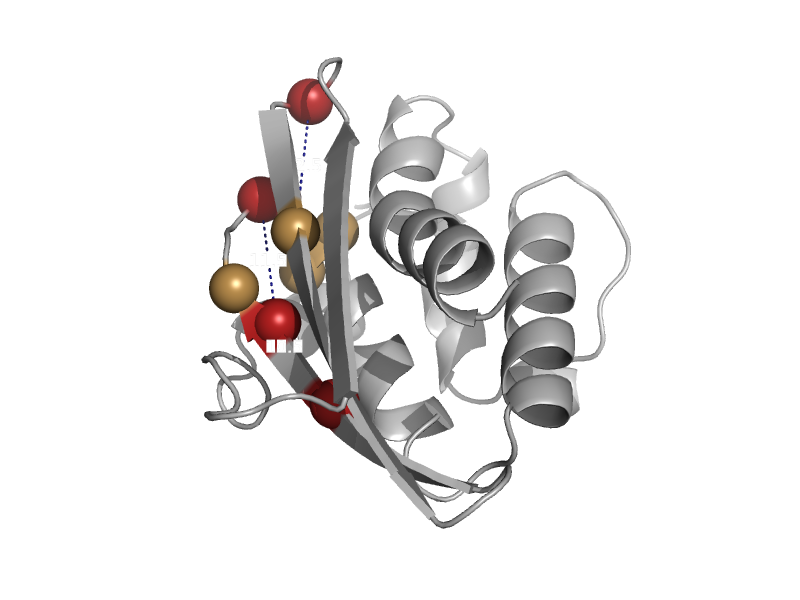 | view | |
| automated matches | automated matches | Methanothrix thermoacetophila [TaxId: 2224] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | S | V | L | D | E | L | Y | R | E | I | L | L | D | H | Y | Q | S | P | R | N | F | G | V | L | P | Q | A | T | K | Q | A | G | G | M | N | P | S | C | G | D | Q | V | E | V | M | V | L | L | E | G | D | T | I | A | D | I | R | F | Q | G | Q | G | C | A | I | S | T | A | S | A | S | L | M | T | E | A | V | K | G | K | K | - | - | V | A | E | A | L | E | L | S | R | K | F | Q | A | M | V | V | E | G | A | P | P | D | P | - | T | L | G | D | L | L | A | L | Q | G | V | A | K | L | P | A | - | R | V | K | C | A | T | L | A | W | H | A | L | E | E | A | L | R | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | A | P | A | F | S | H | L | L | D | E | R | F | T | W | G | G | V | E | L | H | F | D | V | E | K | G | H | I | T | R | A | Q | V | F | T | D | S | L | - | - | N | P | A | P | L | E | A | L | A | G | R | L | Q | G | C | L | Y | R | A | D | M | L | Q | Q | E | C | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | L | L | V | D | F | P | E | Q | - | - | - | - | - | - | - | E | K | E | L | R | E | L | S | A | W | M | A | G | A | V | R | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | M | A | A | L | P | D | K | E | K | L | L | R | N | F | T | R | C | A | N | W | E | E | K | Y | L | Y | I | I | E | L | G | Q | - | - | R | L | A | E | L | N | P | Q | D | R | N | P | - | N | T | I | H | G | - | C | Q | S | Q | V | W | I | V | M | R | R | N | A | N | G | I | I | E | L | Q | G | D | S | D | - | A | A | I | V | K | G | L | M | A | V | V | F | I | L | Y | H | Q | M | T | - | - | A | Q | D | I | V | H | F | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V | R | P | W | F | E | K | M | A | L | A | Q | H | L | T | P | S | R | S | Q | G | L | E | A | M | I | R | A | I | R | A | K | A | A | T | L | S | - | - | - | - |
| N | P | Q | F | A | G | H | P | F | G | T | T | V | - | T | A | E | T | L | R | N | T | F | A | P | L | T | Q | W | E | D | K | Y | R | Q | L | I | M | L | G | K | - | - | Q | L | P | A | L | P | D | E | L | K | A | Q | A | K | E | I | A | G | - | C | E | N | R | V | W | L | G | Y | T | V | A | E | N | G | K | M | H | F | F | G | D | S | E | - | G | R | I | V | R | G | L | L | A | V | L | L | T | A | V | E | G | K | T | - | - | A | A | E | L | Q | A | Q | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | L | A | L | F | D | E | L | G | L | R | A | Q | L | S | A | S | R | S | Q | G | L | N | A | L | S | E | A | I | I | A | A | A | K | Q | V | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | S | P | A | F | N | L | E | R | R | H | R | F | P | I | G | S | I | E | M | K | M | N | V | A | D | G | A | I | Q | E | I | K | I | F | G | D | F | F | - | - | G | L | G | E | I | K | D | V | E | D | I | L | T | G | V | K | Y | D | K | A | S | L | E | E | A | I | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | I | D | V | K | K | Y | F | G | - | - | - | - | - | - | - | N | I | E | K | E | D | L | L | G | L | I | Y | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | A | P | K | F | T | I | K | K | E | E | K | F | K | G | G | I | V | D | A | R | L | T | V | E | K | G | K | I | I | E | L | T | I | Y | G | D | Y | F | - | - | A | K | K | E | T | T | E | I | V | A | A | L | L | G | V | D | Y | Q | Y | S | S | I | W | Q | A | L | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | F | N | F | E | D | Y | F | V | - | - | - | - | - | - | - | N | I | T | K | E | E | F | V | H | L | L | V | D | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | I | D | D | I | Y | N | R | I | L | - | F | A | G | N | M | E | R | I | G | - | L | A | E | P | D | A | V | A | T | V | S | - | - | K | C | G | S | T | V | T | V | Y | L | K | M | R | D | G | V | V | T | D | F | A | H | E | V | - | A | C | A | L | G | Q | A | S | S | S | V | M | A | R | N | V | I | G | A | T | - | - | A | D | E | L | R | A | A | R | D | A | M | Y | R | M | L | K | E | N | G | P | A | P | E | G | R | F | A | D | M | K | Y | F | E | P | V | R | D | Y | - | A | - | R | H | A | S | T | L | L | T | F | D | A | V | A | D | C | I | R | Q | I | E | - | - | - | A |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | I | Y | S | E | F | I | M | D | Y | S | K | L | K | K | F | H | G | K | I | E | N | A | H | K | V | E | E | G | K | N | L | S | C | G | D | E | V | T | L | Y | F | L | F | D | G | D | K | I | V | D | V | K | F | E | G | H | G | C | A | I | S | Q | A | S | T | N | V | M | I | E | Q | I | I | G | K | T | - | - | K | Q | E | A | L | E | M | M | K | N | A | E | N | M | M | L | - | G | K | E | F | D | E | N | V | L | G | P | I | I | N | F | Y | D | V | K | N | Y | P | M | - | R | V | K | C | F | L | L | P | W | K | T | L | E | I | A | L | K | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | I | Q | Q | T | G | - | Y | S | K | K | V | M | E | H | F | M | N | P | R | N | V | G | V | I | D | D | P | D | G | Y | G | K | V | G | N | P | V | C | G | D | L | M | E | I | F | I | K | V | G | D | E | K | I | E | D | I | K | F | R | T | F | G | C | G | A | A | I | A | T | S | S | M | I | T | E | M | A | R | G | K | S | - | - | L | E | E | A | M | R | I | T | R | N | D | V | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | A | L | D | G | L | P | P | - | - | Q | - | K | M | A | C | S | N | L | A | A | D | A | L | H | A | A | I | N | D | Y | L | S | K | K | Q |
