solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1n4ka2 ( 236 ) ggdvvrLfHa----eqekFLTCdehr-----kkqhVFLrttgr-
d1t9fa1 ( 22 ) dedfVtÇysvLkFiNA----ndgSrLHShdvkYgsgsgqqSVTAVk----
d1xzza1 ( 7 ) sfLhigdiCSLYAeg---stnGFISTlg------lvddrCVVqpeagd
d3im6a- ( 13 ) fLrtddeVvLqCtAtihqqkLCLAAegg-------nrlCfLe---st
d3mala- ( 35 ) veITYGSaIKLmHe----ktkfrLHShdvpYgsgSgqqsVTGfp----
bbbbbb bbbb bbbb
60 70 80 90 100
d1n4ka2 ( 270 ) qsatsatsskAlWevevvq-------------------------------
d1t9fa1 ( 64 ) ----nsddinSHWqIfpalna-----------------------------
d1xzza1 ( 46 ) lnnpPkkfrDClFkLcpmnrYsaQkqfwkasttdavllnklhhaAdlekk
d3im6a- ( 53 ) ---sppdlsiCtFvLeqSlsvraLqemlant-------------------
d3mala- ( 75 ) ----gvvdsnSyWiVkpvpgt-----------------------------
333 bbbb
110 120 130 140 150
d1n4ka2 ( 302 ) ------------------lfrFkHlatghyLAAev-------dmvysLvS
d1t9fa1 ( 81 ) ---kçn---rgdaikÇgdkIrLkHlttgtFLhShh-ftAPlskqhqEVSA
d1xzza1 ( 102 ) qnetenrkllgtvIqYgnvIQLlHlkSnkyLTVnkrlpAlleknAmrVtL
d3im6a- ( 86 ) -----------vtLlYghAILLrHsysgmyLCCls--t-----safdVGL
d3mala- ( 92 ) ---tek---qgdaVksgatIrLQHmktrkWLHShl-haSpisg-nlEVSC
bbbbbb bbbbbb bbbbb
160 170 180 190 200
d1n4ka2 ( 358 ) vpeGn--disSiFeldp---------------yvrLrHlctntwVhstni
d1t9fa1 ( 124 ) fgseaeSdtgDDWtViÇn------gdeWleseqFkLrHavtgSyLSLSgq
d1xzza1 ( 152 ) de---aGnegSwFyIqpfyklrsigdsVvigdkVvLnPvnAgqpLhASsh
d3im6a- ( 150 ) qedtt--geaCWWtIhpas---segekVrvgddLILvSMsseryLhlsyg
d3mala- ( 134 ) fgddtnsdtgDhWkLiIegs----gktWkqdqrVrLqHidtsGyLHShdk
b 333 bbbb bbbbbb bbbbbbb
210 220 230
d1n4ka2 ( 405 ) piDkeeekpvmlkIGTsplkedkEaFaivpv
d1t9fa1 ( 168 ) qfgrpih--gqrEVVGTdsitggSaWkVaeGi
d1xzza1 ( 199 ) qLv-dnp--gCneVNSvncn---TSWkIvlf
d3im6a- ( 198 ) ------n--sswhVdAafqq---tlWsVap
d3mala- ( 180 ) kyqriag--gqQEVCGirekkadNIWlAaeGvylplne
b bbbbbb bbbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| IP3 receptor type 1 binding core, domain 1 | MIR domain | Mouse (Mus musculus) [TaxId: 10090] | 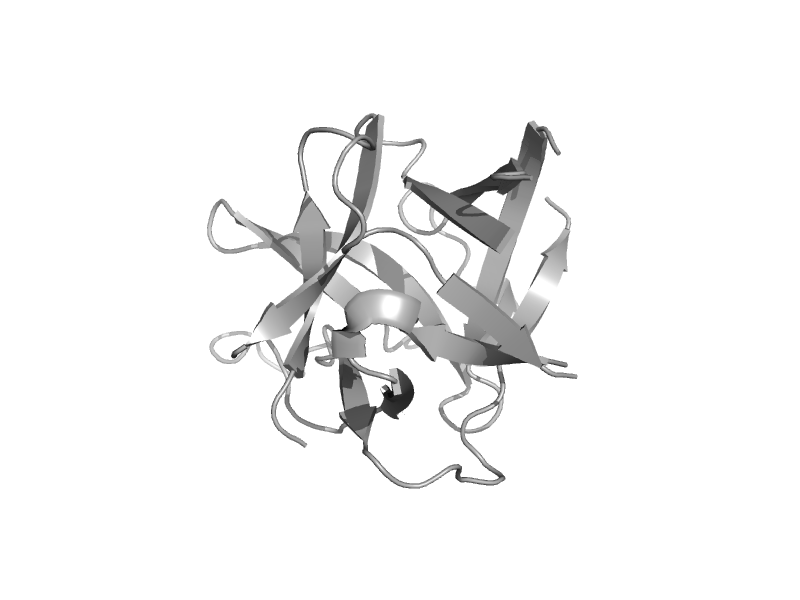 | view | |
| Hypothetical protein R12E2.13 (1D10) | MIR domain | Nematode (Caenorhabditis elegans) [TaxId: 6239] | 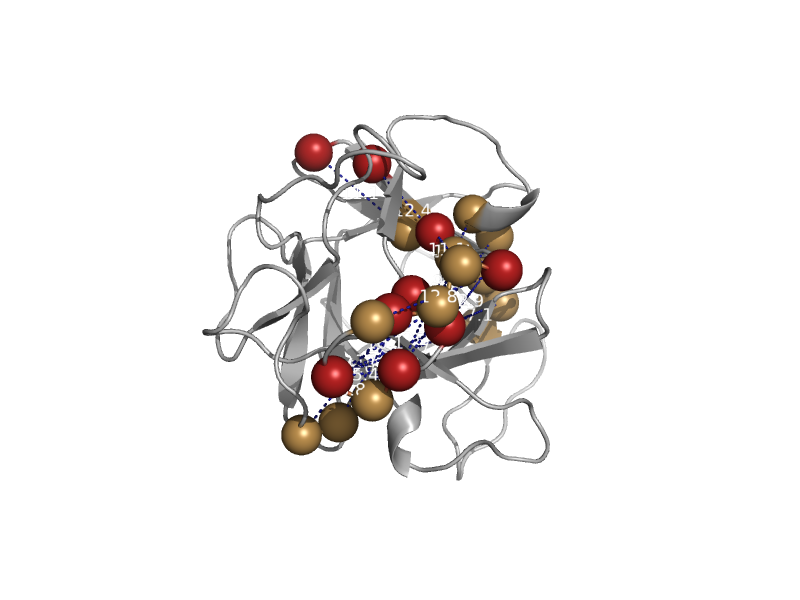 | view | |
| IP3 receptor type 1 ligand binding suppressor domain | Ryanodine receptor N-terminal-like | Mouse (Mus musculus) [TaxId: 10090] | 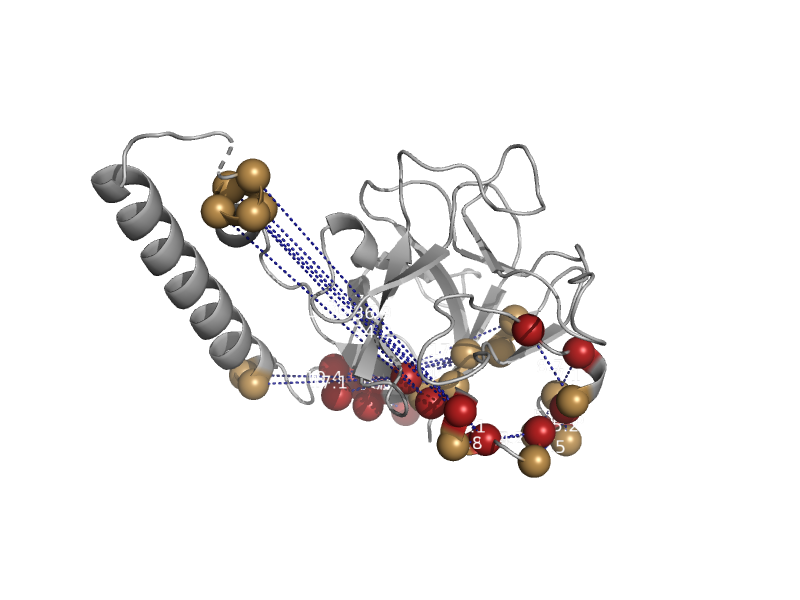 | view | |
| automated matches | Ryanodine receptor N-terminal-like | Mouse (Mus musculus) [TaxId: 10090] | 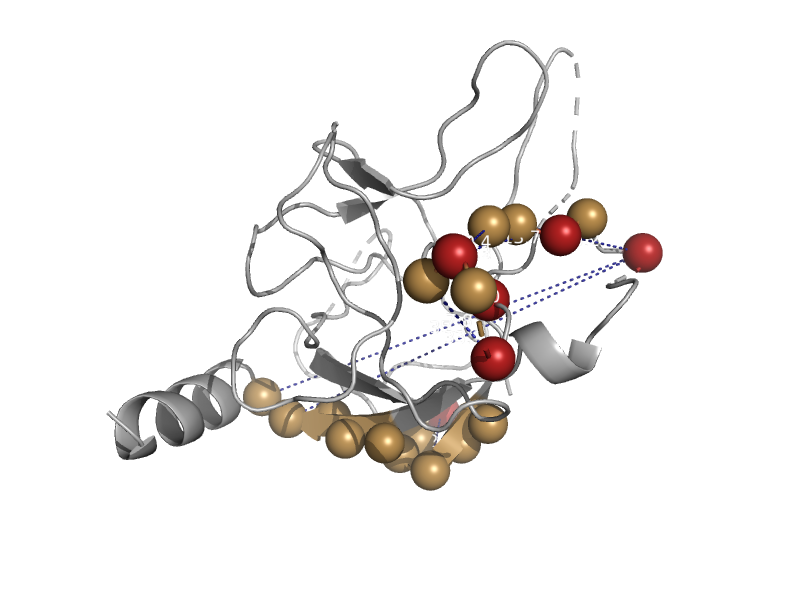 | view | |
| automated matches | automated matches | Thale cress (Arabidopsis thaliana) [TaxId: 3702] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | G | G | D | V | V | R | L | F | H | A | - | - | - | - | E | Q | E | K | F | L | T | C | D | E | H | R | - | - | - | - | - | K | K | Q | H | V | F | L | R | T | T | G | R | - | Q | S | A | T | S | A | T | S | S | K | A | L | W | E | V | E | V | V | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | F | R | F | K | H | L | A | T | G | H | Y | L | A | A | E | V | - | - | - | - | - | - | - | D | M | V | Y | S | L | V | S | V | P | E | G | N | - | - | D | I | S | S | I | F | E | L | D | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | V | R | L | R | H | L | C | T | N | T | W | V | H | S | T | N | I | P | I | D | K | E | E | E | K | P | V | M | L | K | I | G | T | S | P | L | K | E | D | K | E | A | F | A | I | V | P | V | - | - | - | - | - | - | - |
| D | E | D | F | V | T | C | Y | S | V | L | K | F | I | N | A | - | - | - | - | N | D | G | S | R | L | H | S | H | D | V | K | Y | G | S | G | S | G | Q | Q | S | V | T | A | V | K | - | - | - | - | - | - | - | - | N | S | D | D | I | N | S | H | W | Q | I | F | P | A | L | N | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | C | N | - | - | - | R | G | D | A | I | K | C | G | D | K | I | R | L | K | H | L | T | T | G | T | F | L | H | S | H | H | - | F | T | A | P | L | S | K | Q | H | Q | E | V | S | A | F | G | S | E | A | E | S | D | T | G | D | D | W | T | V | I | C | N | - | - | - | - | - | - | G | D | E | W | L | E | S | E | Q | F | K | L | R | H | A | V | T | G | S | Y | L | S | L | S | G | Q | Q | F | G | R | P | I | H | - | - | G | Q | R | E | V | V | G | T | D | S | I | T | G | G | S | A | W | K | V | A | E | G | I | - | - | - | - | - | - |
| - | - | S | F | L | H | I | G | D | I | C | S | L | Y | A | E | G | - | - | - | S | T | N | G | F | I | S | T | L | G | - | - | - | - | - | - | L | V | D | D | R | C | V | V | Q | P | E | A | G | D | L | N | N | P | P | K | K | F | R | D | C | L | F | K | L | C | P | M | N | R | Y | S | A | Q | K | Q | F | W | K | A | S | T | T | D | A | V | L | L | N | K | L | H | H | A | A | D | L | E | K | K | Q | N | E | T | E | N | R | K | L | L | G | T | V | I | Q | Y | G | N | V | I | Q | L | L | H | L | K | S | N | K | Y | L | T | V | N | K | R | L | P | A | L | L | E | K | N | A | M | R | V | T | L | D | E | - | - | - | A | G | N | E | G | S | W | F | Y | I | Q | P | F | Y | K | L | R | S | I | G | D | S | V | V | I | G | D | K | V | V | L | N | P | V | N | A | G | Q | P | L | H | A | S | S | H | Q | L | V | - | D | N | P | - | - | G | C | N | E | V | N | S | V | N | C | N | - | - | - | T | S | W | K | I | V | L | F | - | - | - | - | - | - | - |
| - | - | - | F | L | R | T | D | D | E | V | V | L | Q | C | T | A | T | I | H | Q | Q | K | L | C | L | A | A | E | G | G | - | - | - | - | - | - | - | N | R | L | C | F | L | E | - | - | - | S | T | - | - | - | S | P | P | D | L | S | I | C | T | F | V | L | E | Q | S | L | S | V | R | A | L | Q | E | M | L | A | N | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V | T | L | L | Y | G | H | A | I | L | L | R | H | S | Y | S | G | M | Y | L | C | C | L | S | - | - | T | - | - | - | - | - | S | A | F | D | V | G | L | Q | E | D | T | T | - | - | G | E | A | C | W | W | T | I | H | P | A | S | - | - | - | S | E | G | E | K | V | R | V | G | D | D | L | I | L | V | S | M | S | S | E | R | Y | L | H | L | S | Y | G | - | - | - | - | - | - | N | - | - | S | S | W | H | V | D | A | A | F | Q | Q | - | - | - | T | L | W | S | V | A | P | - | - | - | - | - | - | - | - |
| - | - | V | E | I | T | Y | G | S | A | I | K | L | M | H | E | - | - | - | - | K | T | K | F | R | L | H | S | H | D | V | P | Y | G | S | G | S | G | Q | Q | S | V | T | G | F | P | - | - | - | - | - | - | - | - | G | V | V | D | S | N | S | Y | W | I | V | K | P | V | P | G | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | E | K | - | - | - | Q | G | D | A | V | K | S | G | A | T | I | R | L | Q | H | M | K | T | R | K | W | L | H | S | H | L | - | H | A | S | P | I | S | G | - | N | L | E | V | S | C | F | G | D | D | T | N | S | D | T | G | D | H | W | K | L | I | I | E | G | S | - | - | - | - | G | K | T | W | K | Q | D | Q | R | V | R | L | Q | H | I | D | T | S | G | Y | L | H | S | H | D | K | K | Y | Q | R | I | A | G | - | - | G | Q | Q | E | V | C | G | I | R | E | K | K | A | D | N | I | W | L | A | A | E | G | V | Y | L | P | L | N | E |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1n4ka2 | A | P11881 | GO:0005220 |
| d1n4ka2 | A | P11881 | GO:0005262 |
| d1n4ka2 | A | P11881 | GO:0005783 |
| d1n4ka2 | A | P11881 | GO:0006816 |
| d1n4ka2 | A | P11881 | GO:0016020 |
| d1n4ka2 | A | P11881 | GO:0070588 |
| d1n4ka2 | A | P11881 | GO:0070679 |
