solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1ee8a2 ( 1 ) pelpeVettrrrLrplVlgqtLr-qVvhrdpa---rYr---n-TalAe-g
d1k3xa2 ( 1 ) pegpeIrraAdnLeaaIkgkpLt-dVwFafp-------qLktyqsqLi-g
d1k82a2 ( 1 ) pelpeVetsrrgIephLvgatIl-hAvvrngrLrwpVs---eeIyrLs-d
d1r2za2 ( 2 ) pqlpeVetirrtLlplIvgktIe-dVrIfwpnIIrhprdseaFaarMi-g
d1tdza2 ( 2 ) elpeVetvrreLe-rIvgqkIi-sIeAtyp-MVl--tgfeqLkkeLt-g
d5itqa1 ( 2 ) pegpeLhlasqfVneaCralvFggcVeks-svSrnp-------eVpFeSs
aaaaaaaaaaaaaaa bbbb
60 70 80 90 100
d1ee8a2 ( 42 ) rrIleVdrrgkfLlFaLeg---------gVELVAhLgmtGgFrle----p
d1k3xa2 ( 42 ) qhVthVetrgkALLThFsn---------dlTLYShnqlyGvWrvvdtgep
d1k82a2 ( 46 ) qpVlsVqrrakyLLLeLpe----------gwIIIhLgmsGsLrilpelpp
d1r2za2 ( 50 ) qtVrgLerrgkfLkFlL-d---------rDALIShLrmeGrYavasalpl
d1tdza2 ( 47 ) ktI-gIsrrgkyLiFeIgd---------dfrLIShlrmeG-Yrlatldap
d5itqa1 ( 44 ) aYrIsAsArgkeLrLiLsplpgAqpqqeplaLVFrFgmsGsFqlvpreel
bbbbb bbbbb bbbbb bbbbb
110 120 130 140
d1ee8a2 ( 79 ) tph-trAaLvLeg----rtLyFhDprrfGrLfGVrrgdyreIplLlrl
d1k3xa2 ( 85 ) ttrvlrVkLqTad----kTIlLysAs---dIeMLrpeqltthpfLq-r
d1k82a2 ( 87 ) ekh-DHVDLvMsn---gkvLRYtDprrfGAWlwtelg----hnvlthl
d1r2za2 ( 91 ) eph-ThVVFcFtd---gsELrYrDvrkfGTMhVYakeeAdrrppLa-l
d1tdza2 ( 89 ) ekh-dHLtMkFad----gQLiYaDvrkfGtWelistdqvlpyFlkk-i
d5itqa1 ( 94 ) prh-AHLrFyTappgprlALCFvDirrfGrWdlggkwqp
bbbbbb bbbbb bbbbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| DNA repair protein MutM (Fpg) | N-terminal domain of MutM-like DNA repair proteins | Thermus thermophilus [TaxId: 274] | 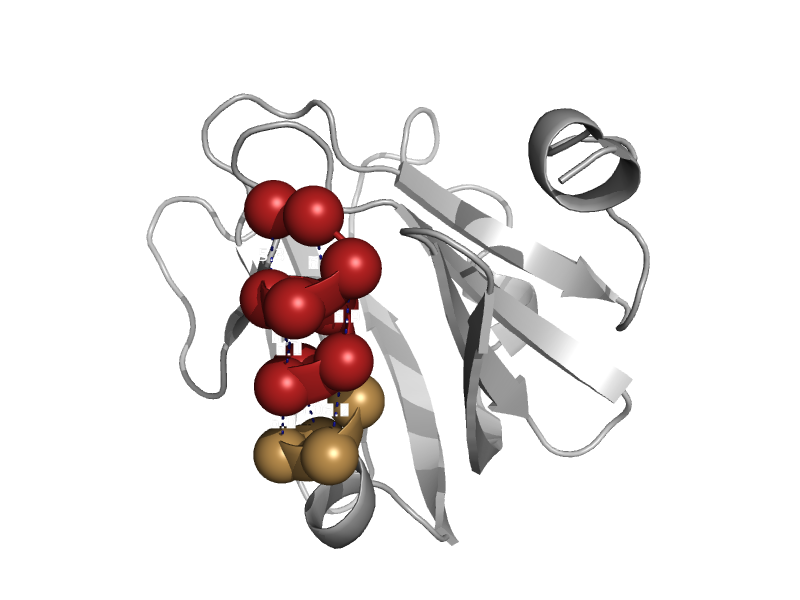 | view | |
| Endonuclease VIII | N-terminal domain of MutM-like DNA repair proteins | Escherichia coli [TaxId: 562] | 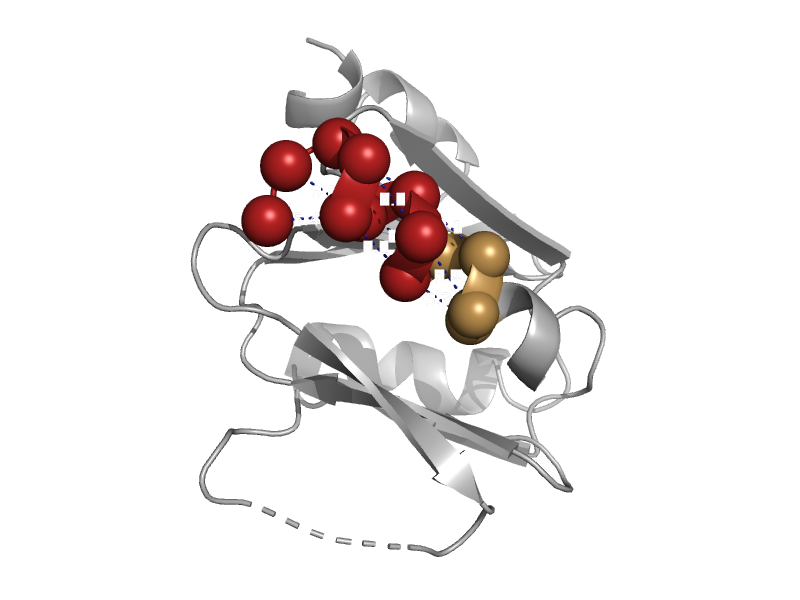 | view | |
| DNA repair protein MutM (Fpg) | N-terminal domain of MutM-like DNA repair proteins | Escherichia coli [TaxId: 562] | 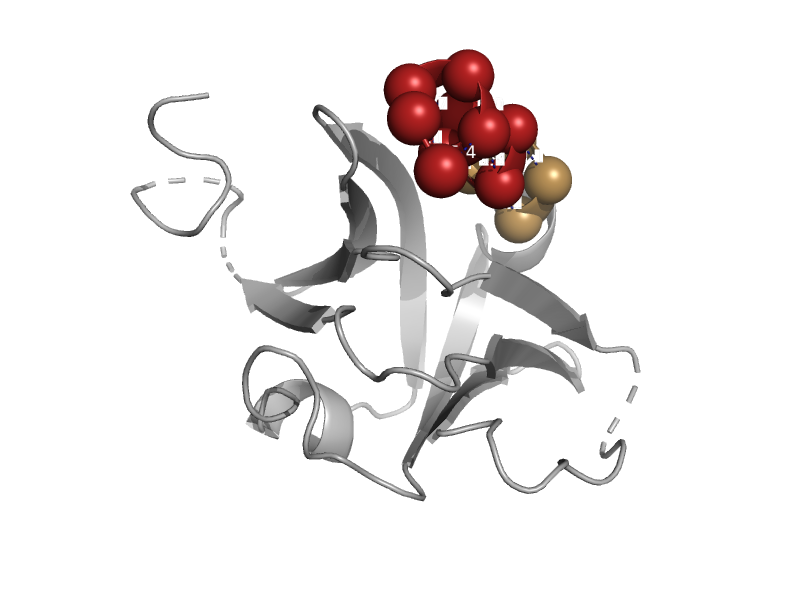 | view | |
| DNA repair protein MutM (Fpg) | N-terminal domain of MutM-like DNA repair proteins | Bacillus stearothermophilus [TaxId: 1422] | 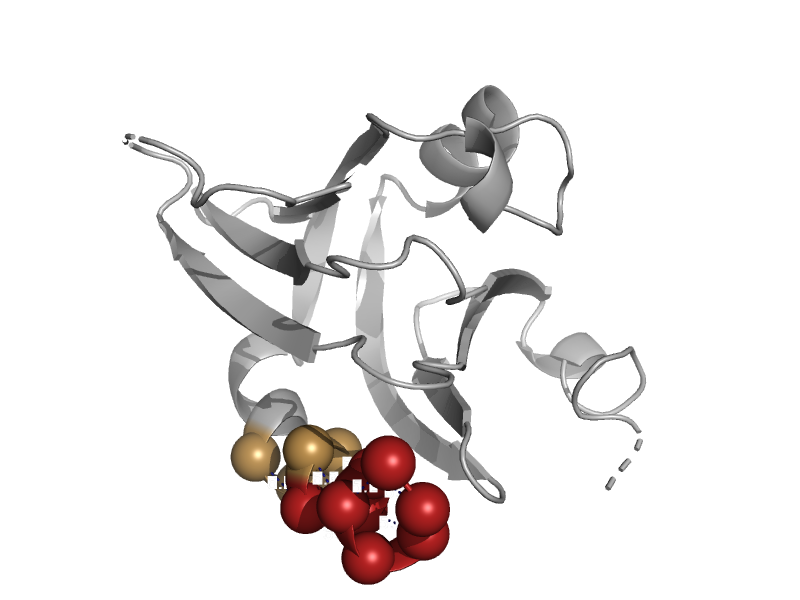 | view | |
| DNA repair protein MutM (Fpg) | N-terminal domain of MutM-like DNA repair proteins | Lactococcus lactis [TaxId: 1358] | 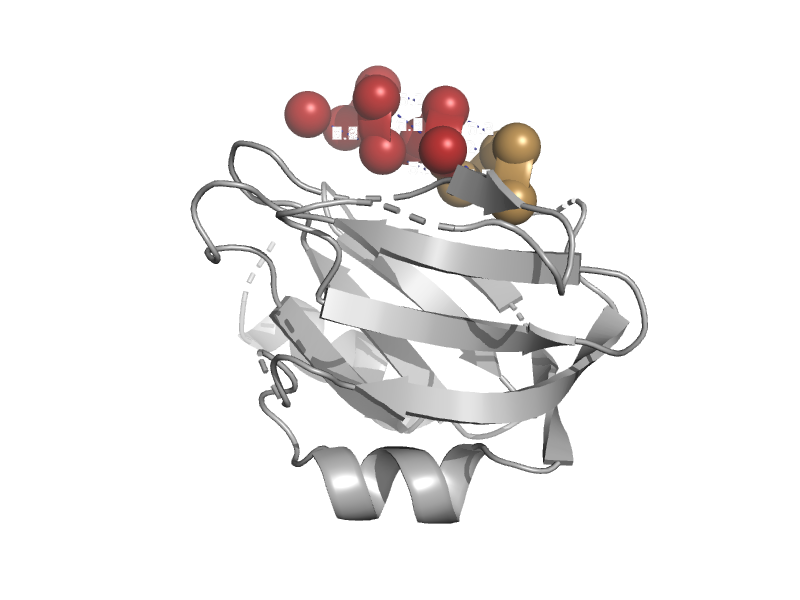 | view | |
| automated matches | N-terminal domain of MutM-like DNA repair proteins | Human (Homo sapiens) [TaxId: 9606] | 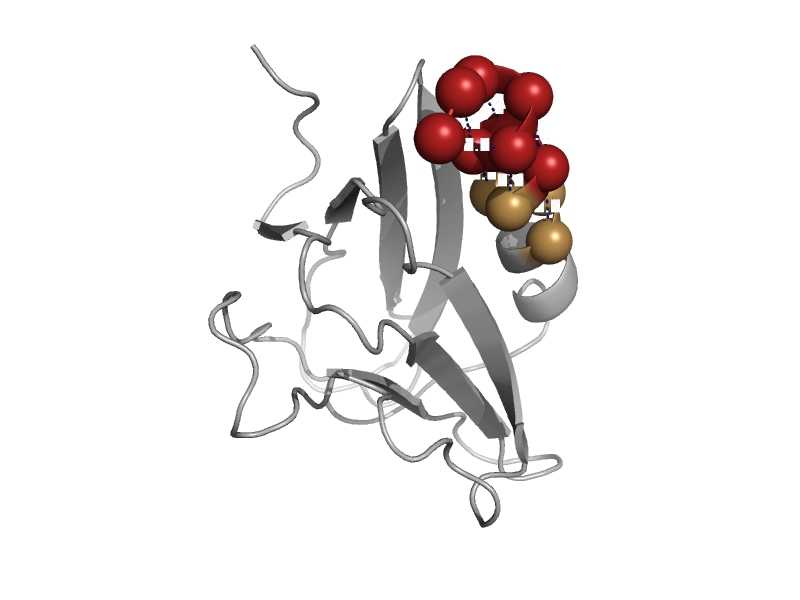 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | E | L | P | E | V | E | T | T | R | R | R | L | R | P | L | V | L | G | Q | T | L | R | - | Q | V | V | H | R | D | P | A | - | - | - | R | Y | R | - | - | - | N | - | T | A | L | A | E | - | G | R | R | I | L | E | V | D | R | R | G | K | F | L | L | F | A | L | E | G | - | - | - | - | - | - | - | - | - | G | V | E | L | V | A | H | L | G | M | T | G | G | F | R | L | E | - | - | - | - | P | T | P | H | - | T | R | A | A | L | V | L | E | G | - | - | - | - | R | T | L | Y | F | H | D | P | R | R | F | G | R | L | F | G | V | R | R | G | D | Y | R | E | I | P | L | L | L | R | L |
| P | E | G | P | E | I | R | R | A | A | D | N | L | E | A | A | I | K | G | K | P | L | T | - | D | V | W | F | A | F | P | - | - | - | - | - | - | - | Q | L | K | T | Y | Q | S | Q | L | I | - | G | Q | H | V | T | H | V | E | T | R | G | K | A | L | L | T | H | F | S | N | - | - | - | - | - | - | - | - | - | D | L | T | L | Y | S | H | N | Q | L | Y | G | V | W | R | V | V | D | T | G | E | P | T | T | R | V | L | R | V | K | L | Q | T | A | D | - | - | - | - | K | T | I | L | L | Y | S | A | S | - | - | - | D | I | E | M | L | R | P | E | Q | L | T | T | H | P | F | L | Q | - | R |
| P | E | L | P | E | V | E | T | S | R | R | G | I | E | P | H | L | V | G | A | T | I | L | - | H | A | V | V | R | N | G | R | L | R | W | P | V | S | - | - | - | E | E | I | Y | R | L | S | - | D | Q | P | V | L | S | V | Q | R | R | A | K | Y | L | L | L | E | L | P | E | - | - | - | - | - | - | - | - | - | - | G | W | I | I | I | H | L | G | M | S | G | S | L | R | I | L | P | E | L | P | P | E | K | H | - | D | H | V | D | L | V | M | S | N | - | - | - | G | K | V | L | R | Y | T | D | P | R | R | F | G | A | W | L | W | T | E | L | G | - | - | - | - | H | N | V | L | T | H | L |
| P | Q | L | P | E | V | E | T | I | R | R | T | L | L | P | L | I | V | G | K | T | I | E | - | D | V | R | I | F | W | P | N | I | I | R | H | P | R | D | S | E | A | F | A | A | R | M | I | - | G | Q | T | V | R | G | L | E | R | R | G | K | F | L | K | F | L | L | - | D | - | - | - | - | - | - | - | - | - | R | D | A | L | I | S | H | L | R | M | E | G | R | Y | A | V | A | S | A | L | P | L | E | P | H | - | T | H | V | V | F | C | F | T | D | - | - | - | G | S | E | L | R | Y | R | D | V | R | K | F | G | T | M | H | V | Y | A | K | E | E | A | D | R | R | P | P | L | A | - | L |
| - | E | L | P | E | V | E | T | V | R | R | E | L | E | - | R | I | V | G | Q | K | I | I | - | S | I | E | A | T | Y | P | - | M | V | L | - | - | T | G | F | E | Q | L | K | K | E | L | T | - | G | K | T | I | - | G | I | S | R | R | G | K | Y | L | I | F | E | I | G | D | - | - | - | - | - | - | - | - | - | D | F | R | L | I | S | H | L | R | M | E | G | - | Y | R | L | A | T | L | D | A | P | E | K | H | - | D | H | L | T | M | K | F | A | D | - | - | - | - | G | Q | L | I | Y | A | D | V | R | K | F | G | T | W | E | L | I | S | T | D | Q | V | L | P | Y | F | L | K | K | - | I |
| P | E | G | P | E | L | H | L | A | S | Q | F | V | N | E | A | C | R | A | L | V | F | G | G | C | V | E | K | S | - | S | V | S | R | N | P | - | - | - | - | - | - | - | E | V | P | F | E | S | S | A | Y | R | I | S | A | S | A | R | G | K | E | L | R | L | I | L | S | P | L | P | G | A | Q | P | Q | Q | E | P | L | A | L | V | F | R | F | G | M | S | G | S | F | Q | L | V | P | R | E | E | L | P | R | H | - | A | H | L | R | F | Y | T | A | P | P | G | P | R | L | A | L | C | F | V | D | I | R | R | F | G | R | W | D | L | G | G | K | W | Q | P | - | - | - | - | - | - | - | - | - |
