solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1pp9h- ( 13 ) lvdplttvreqÇeqlekCvkArerlelÇdervssrs--------qteedç
d3cx5f- ( 74 ) vtdqledlrehfknteeGkalvhhyeeÇaervkiqqqqpgyadlehkedç
aaaaaaaaa aaaaaaaaaaaaaaaaaa
60 70
d1pp9h- ( 55 ) teelldflhardhçvahklfnslk
d3cx5f- ( 124 ) veeffhlqhyLdtaTaprlfdklk
aaaaaaaaaaaaaaaaaa 333
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Non-heme 11 kDa protein of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) | Non-heme 11 kDa protein of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) | Cow (Bos taurus) [TaxId: 9913] | 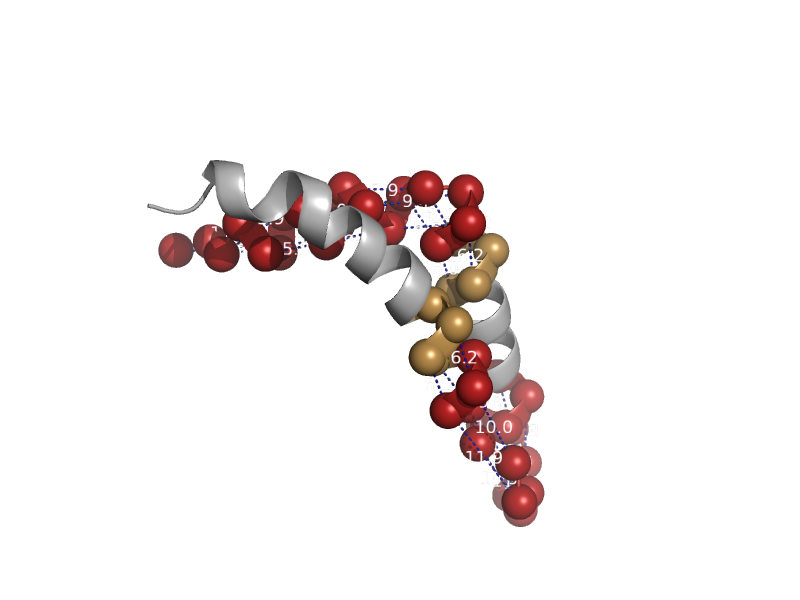 | view | |
| automated matches | Non-heme 11 kDa protein of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) | Saccharomyces cerevisiae [TaxId: 764097] | 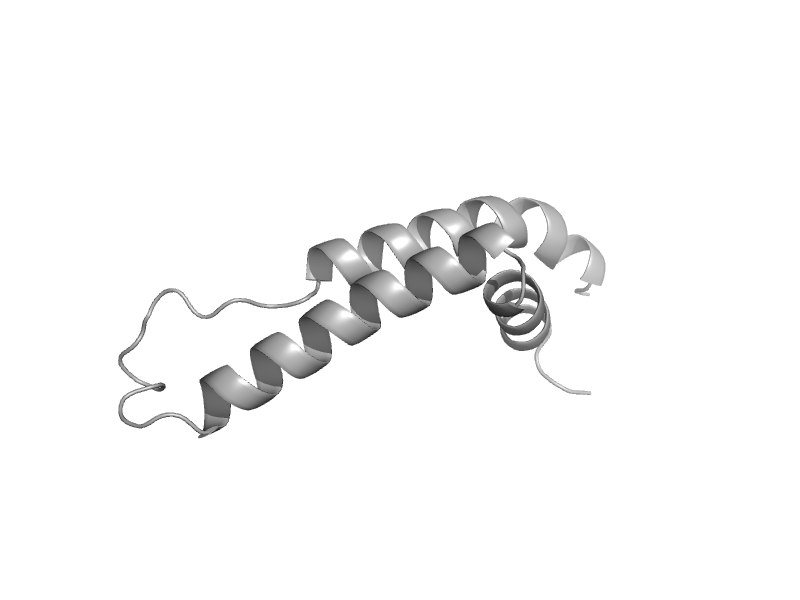 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L | V | D | P | L | T | T | V | R | E | Q | C | E | Q | L | E | K | C | V | K | A | R | E | R | L | E | L | C | D | E | R | V | S | S | R | S | - | - | - | - | - | - | - | - | Q | T | E | E | D | C | T | E | E | L | L | D | F | L | H | A | R | D | H | C | V | A | H | K | L | F | N | S | L | K |
| V | T | D | Q | L | E | D | L | R | E | H | F | K | N | T | E | E | G | K | A | L | V | H | H | Y | E | E | C | A | E | R | V | K | I | Q | Q | Q | Q | P | G | Y | A | D | L | E | H | K | E | D | C | V | E | E | F | F | H | L | Q | H | Y | L | D | T | A | T | A | P | R | L | F | D | K | L | K |
