solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1k3ra2 ( 1 )
d1v2xa- ( 1 )
d2v3ka- ( 25 ) pqappvLtSkdkitkRIVVLAAslethk-------------------iyv
d3aiaa1 ( 2 ) reFIFkAnkTiTssdInlkdLpg-------------
d3bbda1 ( 2 ) tynIILAkSAlElIpeeIknk--IrksrvYk----yd
d3o7ba- ( 1 ) mgSAFVFLEASLelIpqkIrghpaVradairgkrpekI
60 70 80 90 100
d1k3ra2 ( 1 ) mnrVdLSLFIP
d1v2xa- ( 1 ) mrertearrrrieevLr----rRQpdLTVLLe
d2v3ka- ( 67 ) LlncddhqglLkkgrdise-arPdiTHqCLltLldspINkagkLQVYIqT
d3aiaa1 ( 25 ) ------------------sCgrLdLLCrCVsdAFflshdirrdVvFYAVL
d3bbda1 ( 33 ) ILDsnyhykA-ekLkdke-RGrPdIIhiSLlnILdSpINhekkLnIyIHT
d3o7ba- ( 40 ) LLddskHhtAMksLefreKrGrPdiVHqCLllLLdSp---lrdFeVyVhT
aaaaaaaaa bbbbbb
110 120 130 140 150
d1k3ra2 ( 12 ) DSLTaetg----------dlkikTykVVlIArAAsifgVkrIVIYhddad
d1v2xa- ( 29 ) nVh-------------------kphNLSaILrtCdaVgVleAHAv-----
d2v3ka- ( 117 ) sr-----giLIeVnptVr-Iprtfkrfsgl-vqllhk--lsirsv-----
d3aiaa1 ( 57 ) ygqpnppVCIkFvGseLkkVspdernIAifIkkALkkFeeld--------
d3bbda1 ( 83 ) yd-----dkVlkInpeTr-LprnyfrfLgv-ekvLkg--ern--------
d3o7ba- ( 87 ) Ln-----geIIwVnreTr-LprnynrFvglMEkLFee--rritag-----
aaaaaa aaaa
160 170 180 190 200
d1k3ra2 ( 52 ) gearFIrdILtyMdtpqylrrkvfpimreLkhVgiLpplr------tgYe
d1v2xa- ( 55 ) --nPTgg---------------------------vptfnetsggShkwVy
d2v3ka- ( 154 ) --------------------------------------------nseekl
d3aiaa1 ( 99 ) --eeqrk---------------------------dwnqStp------GIy
d3bbda1 ( 117 ) ------------------------------------------------hl
d3o7ba- ( 124 ) ----------------------------------------------dttl
b
210 220 230 240 250
d1k3ra2 ( 167 ) VldtrrnLaeSlktv--gAdvvVAtsr--naspItsildeVktrMrgare
d1v2xa- ( 76 ) LrvHp-dLheAFrfLkergftVyAtalredArdFreV--dYtk------p
d2v3ka- ( 160 ) LkviknpItdhLPt----kCrkVTLsf--dapvi--rVqdyIekLdddeS
d3aiaa1 ( 114 ) Vrrl--gFrnLVleKleegknIYYLhm--ngedVenV--dI-e------n
d3bbda1 ( 119 ) ik-eektLedLLnei---nAkkIAItk--tgklt--hpklLke-----yd
d3o7ba- ( 128 ) IefkdvgLrdiVr-----gRdVLLFre--kGgrf--efseLLd----gdV
bb b aaaa bbbbb
260 270 280 290 300
d1k3ra2 ( 213 ) AAILFgGpykgLpe---IdAd--IwVnTlpgqctetvrTeeaVlaTLsvF
d1v2xa- ( 117 ) TAVLFGAekwgVseeAlalAd--gaIkipmlgmvqslnvsvaAavILfeA
d2v3ka- ( 202 ) ICVFVgAargkdn-fAdeyV----dekVGLs-nyp-lsasvaCskFchGa
d3aiaa1 ( 151 ) pVFIIgdhi-gIgeeDerfLdeikAkrIsLs-ple-lhANhcItiIHnvl
d3bbda1 ( 158 ) TFIigGfpygkLkInkekVf--gdikeISiy-nkg-l-AwtvCgiICysL
d3o7ba- ( 165 ) AVCIgAfphgdffeeTlreL--gefkeVsLg-tes-ytslyvTsrVlceY
bbbbb bb aaaaaaaaaaaa
310 320
d1k3ra2 ( 258 ) nmLtq
d1v2xa- ( 165 ) qrqRlkaglydrprLdpelyqkVladW
d2v3ka- ( 246 ) edAwni
d3aiaa1 ( 198 ) dkk
d3bbda1 ( 204 ) sf
d3o7ba- ( 211 ) ervra
aa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Hypothetical protein MTH1 (MT0001), dimerisation domain | Hypothetical protein MTH1 (MT0001), dimerisation domain | Methanobacterium thermoautotrophicum [TaxId: 145262] | 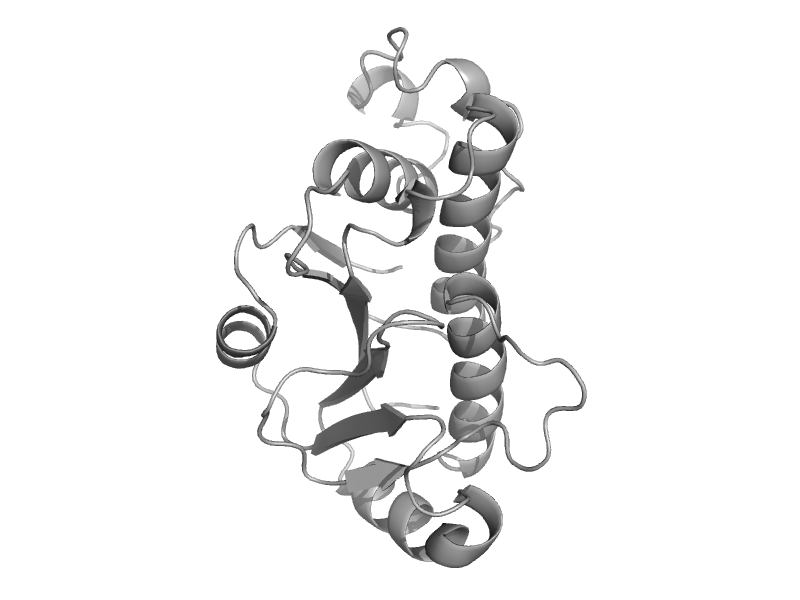 | view | |
| tRNA (Gm18) methyltransferase TrmH | SpoU-like RNA 2'-O ribose methyltransferase | Thermus thermophilus [TaxId: 274] | 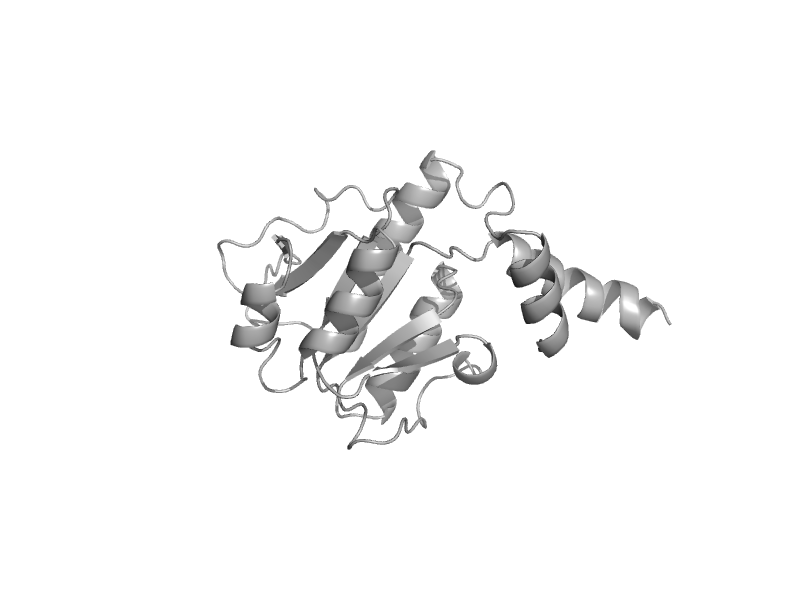 | view | |
| Essential for mitotic growth 1, EMG1 | EMG1/NEP1-like | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 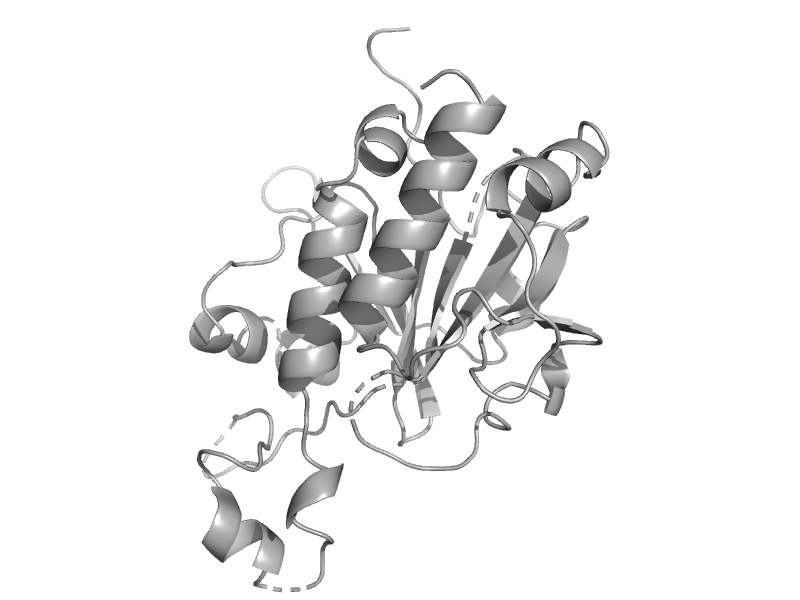 | view | |
| automated matches | automated matches | Methanocaldococcus jannaschii [TaxId: 2190] | 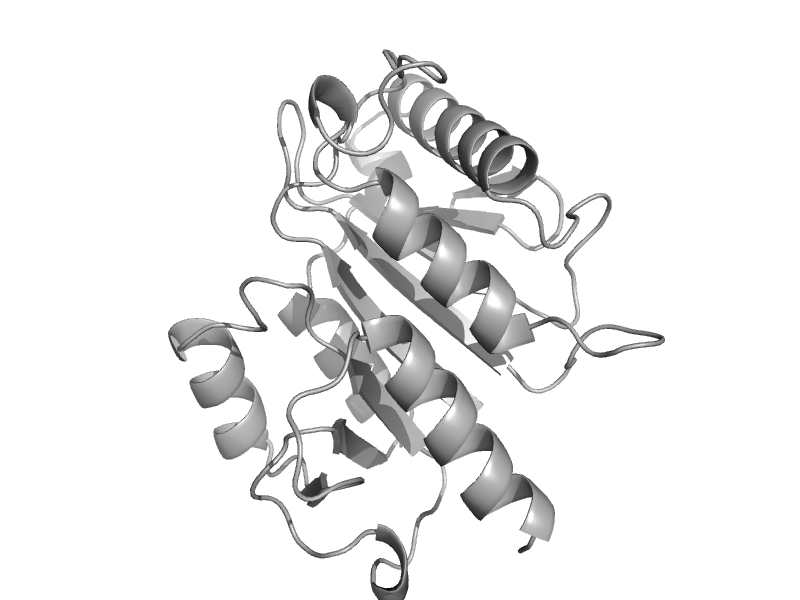 | view | |
| Ribosome biogenesis protein NEP1 | EMG1/NEP1-like | Methanococcus jannaschii [TaxId: 2190] | 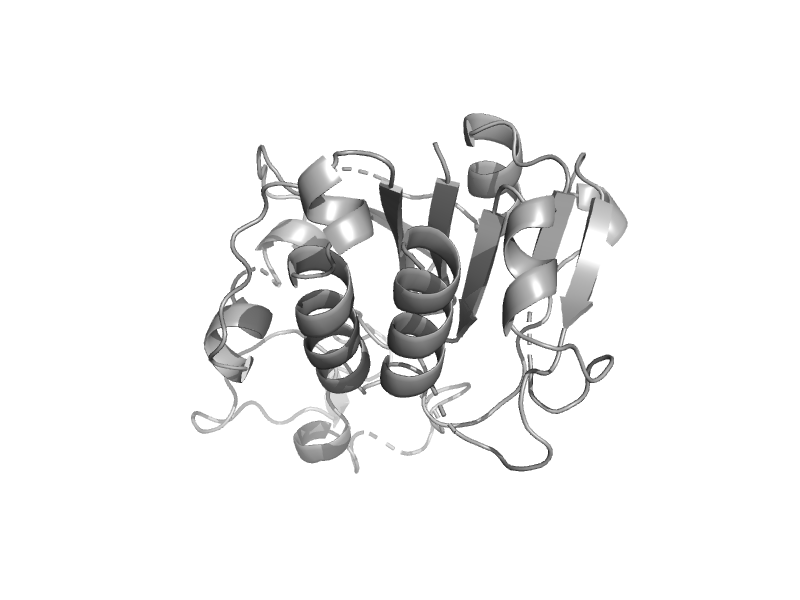 | view | |
| automated matches | automated matches | Archaeoglobus fulgidus [TaxId: 2234] | 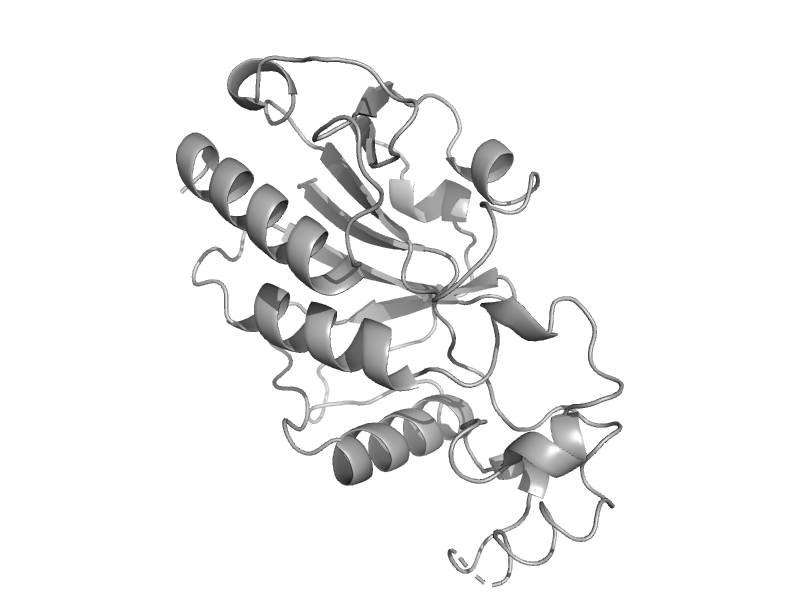 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | N | R | V | D | L | S | L | F | I | P | D | S | L | T | A | E | T | G | - | - | - | - | - | - | - | - | - | - | D | L | K | I | K | T | Y | K | V | V | L | I | A | R | A | A | S | I | F | G | V | K | R | I | V | I | Y | H | D | D | A | D | G | E | A | R | F | I | R | D | I | L | T | Y | M | D | T | P | Q | Y | L | R | R | K | V | F | P | I | M | R | E | L | K | H | V | G | I | L | P | P | L | R | - | - | - | - | - | - | T | G | Y | E | V | L | D | T | R | R | N | L | A | E | S | L | K | T | V | - | - | G | A | D | V | V | V | A | T | S | R | - | - | N | A | S | P | I | T | S | I | L | D | E | V | K | T | R | M | R | G | A | R | E | A | A | I | L | F | G | G | P | Y | K | G | L | P | E | - | - | - | I | D | A | D | - | - | I | W | V | N | T | L | P | G | Q | C | T | E | T | V | R | T | E | E | A | V | L | A | T | L | S | V | F | N | M | L | T | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | R | E | R | T | E | A | R | R | R | R | I | E | E | V | L | R | - | - | - | - | R | R | Q | P | D | L | T | V | L | L | E | N | V | H | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | P | H | N | L | S | A | I | L | R | T | C | D | A | V | G | V | L | E | A | H | A | V | - | - | - | - | - | - | - | N | P | T | G | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V | P | T | F | N | E | T | S | G | G | S | H | K | W | V | Y | L | R | V | H | P | - | D | L | H | E | A | F | R | F | L | K | E | R | G | F | T | V | Y | A | T | A | L | R | E | D | A | R | D | F | R | E | V | - | - | D | Y | T | K | - | - | - | - | - | - | P | T | A | V | L | F | G | A | E | K | W | G | V | S | E | E | A | L | A | L | A | D | - | - | G | A | I | K | I | P | M | L | G | M | V | Q | S | L | N | V | S | V | A | A | A | V | I | L | F | E | A | Q | R | Q | R | L | K | A | G | L | Y | D | R | P | R | L | D | P | E | L | Y | Q | K | V | L | A | D | W |
| P | Q | A | P | P | V | L | T | S | K | D | K | I | T | K | R | I | V | V | L | A | A | S | L | E | T | H | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | I | Y | V | L | L | N | C | D | D | H | Q | G | L | L | K | K | G | R | D | I | S | E | - | A | R | P | D | I | T | H | Q | C | L | L | T | L | L | D | S | P | I | N | K | A | G | K | L | Q | V | Y | I | Q | T | S | R | - | - | - | - | - | G | I | L | I | E | V | N | P | T | V | R | - | I | P | R | T | F | K | R | F | S | G | L | - | V | Q | L | L | H | K | - | - | L | S | I | R | S | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | S | E | E | K | L | L | K | V | I | K | N | P | I | T | D | H | L | P | T | - | - | - | - | K | C | R | K | V | T | L | S | F | - | - | D | A | P | V | I | - | - | R | V | Q | D | Y | I | E | K | L | D | D | D | E | S | I | C | V | F | V | G | A | A | R | G | K | D | N | - | F | A | D | E | Y | V | - | - | - | - | D | E | K | V | G | L | S | - | N | Y | P | - | L | S | A | S | V | A | C | S | K | F | C | H | G | A | E | D | A | W | N | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | E | F | I | F | K | A | N | K | T | I | T | S | S | D | I | N | L | K | D | L | P | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | C | G | R | L | D | L | L | C | R | C | V | S | D | A | F | F | L | S | H | D | I | R | R | D | V | V | F | Y | A | V | L | Y | G | Q | P | N | P | P | V | C | I | K | F | V | G | S | E | L | K | K | V | S | P | D | E | R | N | I | A | I | F | I | K | K | A | L | K | K | F | E | E | L | D | - | - | - | - | - | - | - | - | - | - | E | E | Q | R | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | W | N | Q | S | T | P | - | - | - | - | - | - | G | I | Y | V | R | R | L | - | - | G | F | R | N | L | V | L | E | K | L | E | E | G | K | N | I | Y | Y | L | H | M | - | - | N | G | E | D | V | E | N | V | - | - | D | I | - | E | - | - | - | - | - | - | N | P | V | F | I | I | G | D | H | I | - | G | I | G | E | E | D | E | R | F | L | D | E | I | K | A | K | R | I | S | L | S | - | P | L | E | - | L | H | A | N | H | C | I | T | I | I | H | N | V | L | D | K | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | T | Y | N | I | I | L | A | K | S | A | L | E | L | I | P | E | E | I | K | N | K | - | - | I | R | K | S | R | V | Y | K | - | - | - | - | Y | D | I | L | D | S | N | Y | H | Y | K | A | - | E | K | L | K | D | K | E | - | R | G | R | P | D | I | I | H | I | S | L | L | N | I | L | D | S | P | I | N | H | E | K | K | L | N | I | Y | I | H | T | Y | D | - | - | - | - | - | D | K | V | L | K | I | N | P | E | T | R | - | L | P | R | N | Y | F | R | F | L | G | V | - | E | K | V | L | K | G | - | - | E | R | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | H | L | I | K | - | E | E | K | T | L | E | D | L | L | N | E | I | - | - | - | N | A | K | K | I | A | I | T | K | - | - | T | G | K | L | T | - | - | H | P | K | L | L | K | E | - | - | - | - | - | Y | D | T | F | I | I | G | G | F | P | Y | G | K | L | K | I | N | K | E | K | V | F | - | - | G | D | I | K | E | I | S | I | Y | - | N | K | G | - | L | - | A | W | T | V | C | G | I | I | C | Y | S | L | S | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | M | G | S | A | F | V | F | L | E | A | S | L | E | L | I | P | Q | K | I | R | G | H | P | A | V | R | A | D | A | I | R | G | K | R | P | E | K | I | L | L | D | D | S | K | H | H | T | A | M | K | S | L | E | F | R | E | K | R | G | R | P | D | I | V | H | Q | C | L | L | L | L | L | D | S | P | - | - | - | L | R | D | F | E | V | Y | V | H | T | L | N | - | - | - | - | - | G | E | I | I | W | V | N | R | E | T | R | - | L | P | R | N | Y | N | R | F | V | G | L | M | E | K | L | F | E | E | - | - | R | R | I | T | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | T | T | L | I | E | F | K | D | V | G | L | R | D | I | V | R | - | - | - | - | - | G | R | D | V | L | L | F | R | E | - | - | K | G | G | R | F | - | - | E | F | S | E | L | L | D | - | - | - | - | G | D | V | A | V | C | I | G | A | F | P | H | G | D | F | F | E | E | T | L | R | E | L | - | - | G | E | F | K | E | V | S | L | G | - | T | E | S | - | Y | T | S | L | Y | V | T | S | R | V | L | C | E | Y | E | R | V | R | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
