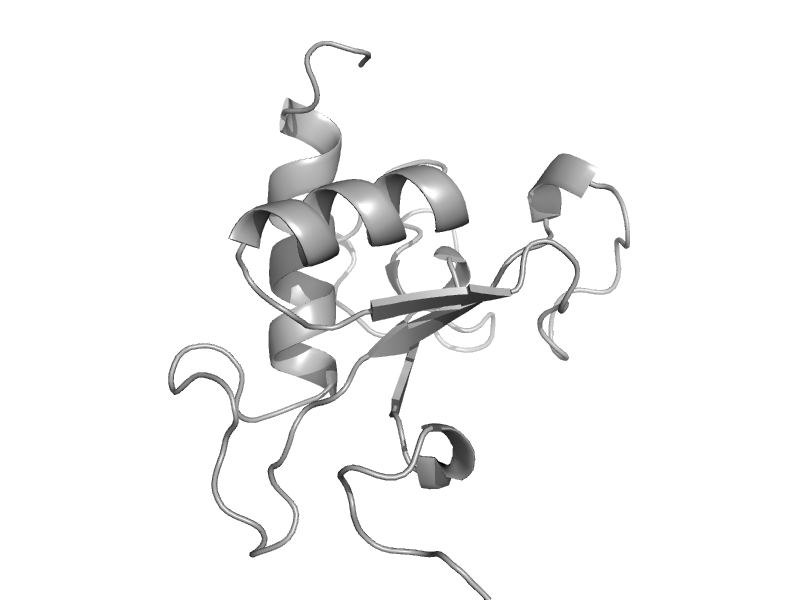
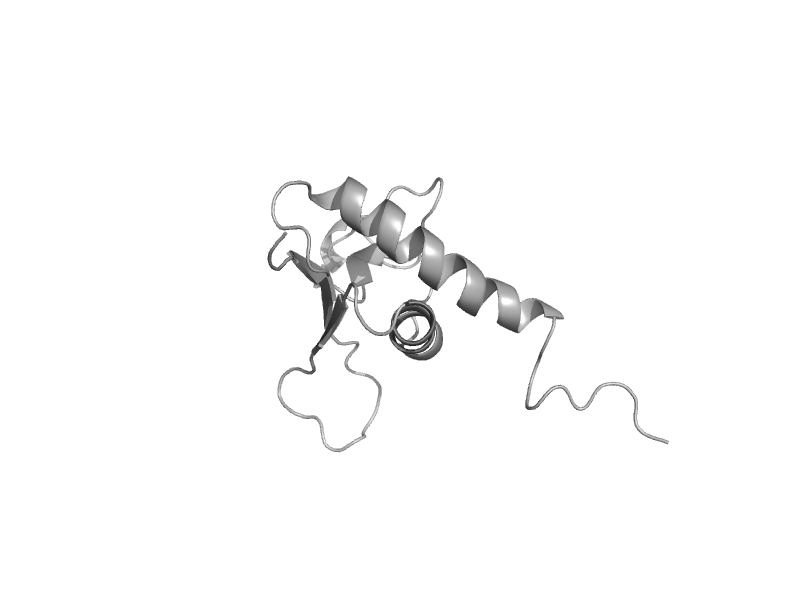
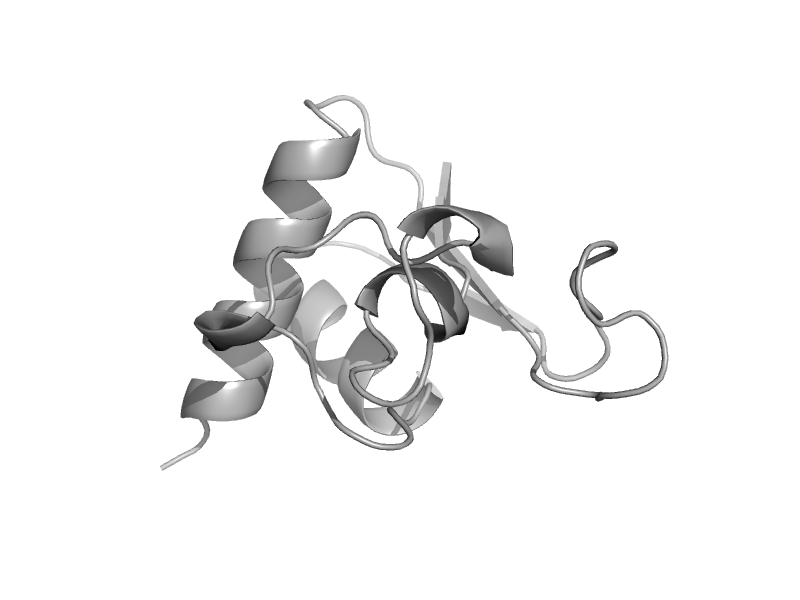
solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1jida- ( 5 ) aarspadqdrFicIypayLnnkktiaeGrrIpiskAVenPtAteIqdVCs
d1kvna- ( 1 ) mkesvvwtvnlDskksraegrriprrfAvpnvklhElveAsk
d1lnga- ( 1 ) miIwpSyIdkkksrreGRkVpeelAiekPsLkdIekALk
d3dlua1 ( 2 ) grfvVwpseLdsrlsr-ygriVprsiAv-sPrV-eIvrAAe
bbb 3333 aaaaaaaaa
60 70 80 90 100
d1jida- ( 55 ) avgLnV-fleknkmysrewnrdvqyrGrVrVqlkqedgslclvqFpsrks
d1kvna- ( 43 ) elglkF-raeekkypks----wweeggrvvvekr-----------gtktk
d1lnga- ( 40 ) klgleP-kiyrdkryprQh---weicGCVeVdyk-----------gnklq
d3dlua1 ( 43 ) elkFkvirveedklnpl---rt---fGmIvLeSpy-----------gksk
a b bbb bbbb aaa
110 120
d1jida- ( 104 ) VMlyAAemIpklktr
d1kvna- ( 77 ) lMielArkiaeireqkreqkkdkkkkkk
d1lnga- ( 75 ) LLkeICkiikgkn
d3dlua1 ( 84 ) SLklIAqkIreFrrrsa
aaaaaaaaa
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | A | R | S | P | A | D | Q | D | R | F | I | C | I | Y | P | A | Y | L | N | N | K | K | T | I | A | E | G | R | R | I | P | I | S | K | A | V | E | N | P | T | A | T | E | I | Q | D | V | C | S | A | V | G | L | N | V | - | F | L | E | K | N | K | M | Y | S | R | E | W | N | R | D | V | Q | Y | R | G | R | V | R | V | Q | L | K | Q | E | D | G | S | L | C | L | V | Q | F | P | S | R | K | S | V | M | L | Y | A | A | E | M | I | P | K | L | K | T | R | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | M | K | E | S | V | V | W | T | V | N | L | D | S | K | K | S | R | A | E | G | R | R | I | P | R | R | F | A | V | P | N | V | K | L | H | E | L | V | E | A | S | K | E | L | G | L | K | F | - | R | A | E | E | K | K | Y | P | K | S | - | - | - | - | W | W | E | E | G | G | R | V | V | V | E | K | R | - | - | - | - | - | - | - | - | - | - | - | G | T | K | T | K | L | M | I | E | L | A | R | K | I | A | E | I | R | E | Q | K | R | E | Q | K | K | D | K | K | K | K | K | K |
| - | - | - | - | - | - | - | - | - | - | - | M | I | I | W | P | S | Y | I | D | K | K | K | S | R | R | E | G | R | K | V | P | E | E | L | A | I | E | K | P | S | L | K | D | I | E | K | A | L | K | K | L | G | L | E | P | - | K | I | Y | R | D | K | R | Y | P | R | Q | H | - | - | - | W | E | I | C | G | C | V | E | V | D | Y | K | - | - | - | - | - | - | - | - | - | - | - | G | N | K | L | Q | L | L | K | E | I | C | K | I | I | K | G | K | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | G | R | F | V | V | W | P | S | E | L | D | S | R | L | S | R | - | Y | G | R | I | V | P | R | S | I | A | V | - | S | P | R | V | - | E | I | V | R | A | A | E | E | L | K | F | K | V | I | R | V | E | E | D | K | L | N | P | L | - | - | - | R | T | - | - | - | F | G | M | I | V | L | E | S | P | Y | - | - | - | - | - | - | - | - | - | - | - | G | K | S | K | S | L | K | L | I | A | Q | K | I | R | E | F | R | R | R | S | A | - | - | - | - | - | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1jida_ | A | P09132 | GO:0006614 |
| d1jida_ | A | P09132 | GO:0008312 |
| d1jida_ | A | P09132 | GO:0048500 |
| d1jida_ | A | P09132 | GO:0006614 |
| d1jida_ | A | P09132 | GO:0008312 |
| d1jida_ | A | P09132 | GO:0048500 |
| d1kvna_ | A | O29010 | GO:0003723 |
| d1kvna_ | A | O29010 | GO:0005737 |
| d1kvna_ | A | O29010 | GO:0006612 |
| d1kvna_ | A | O29010 | GO:0006614 |
| d1kvna_ | A | O29010 | GO:0006617 |
| d1kvna_ | A | O29010 | GO:0008312 |
| d1kvna_ | A | O29010 | GO:0048500 |
| d1kvna_ | A | O29010 | GO:0003723 |
| d1kvna_ | A | O29010 | GO:0005737 |
| d1kvna_ | A | O29010 | GO:0006612 |
| d1kvna_ | A | O29010 | GO:0006614 |
| d1kvna_ | A | O29010 | GO:0006617 |
| d1kvna_ | A | O29010 | GO:0008312 |
| d1kvna_ | A | O29010 | GO:0048500 |
| d1lnga_ | A | Q58440 | GO:0003723 |
| d1lnga_ | A | Q58440 | GO:0005737 |
| d1lnga_ | A | Q58440 | GO:0006612 |
| d1lnga_ | A | Q58440 | GO:0006614 |
| d1lnga_ | A | Q58440 | GO:0008312 |
| d1lnga_ | A | Q58440 | GO:0048500 |
| d1lnga_ | A | Q58440 | GO:0003723 |
| d1lnga_ | A | Q58440 | GO:0005737 |
| d1lnga_ | A | Q58440 | GO:0006612 |
| d1lnga_ | A | Q58440 | GO:0006614 |
| d1lnga_ | A | Q58440 | GO:0008312 |
| d1lnga_ | A | Q58440 | GO:0048500 |