solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1ii2a2 ( 2 ) pPtihrnllspe
d1j3ba2 ( 2 ) q----rLealg--Ihpkkrvfwntvspv
d2olra2 ( 6 ) gLtpqeLeayg--IsdVhdivynPsydl
d4gmua1 ( 3 ) pqlhngldfsakViqgsLdsLpqeVrkfVegnAqlCqPeyIhICdGseee
bbb aaa
60 70 80 90 100
d1ii2a2 ( 14 ) LvqwAlk-----iedS-ltar--gaLavmsya-ktgrspldkRIVdTddV
d1j3ba2 ( 24 ) LvehTll-----rgegllahh--gpLvvdttp-ytgrspkDkfVVrepeV
d2olra2 ( 32 ) LyqeEldpsltgyerGvltnl--gaVavdtGi-ftgrspkDkYIVrddtT
d4gmua1 ( 53 ) ygrlLahMqe--egvIrklkkydncwlaltDprdvariesktViI-tqeq
aaaaaaa bb bb 333bbbb
110 120 130 140 150
d1ii2a2 ( 57 ) r-nvdwg------vnmkLseeSFarVrkiAkefLdtrehLFVvDCFAghd
d1j3ba2 ( 66 ) egeiwwg-----evnqpfapeaFeaLYqrVVqyLser-dLYVQdLyagad
d2olra2 ( 79 ) rdtfwwad-ggkndnkpLspetWqhLkglVtrqLsgk-rLFVVDAFcgan
d4gmua1 ( 100 ) rdtVpipksgqsqlgrwmseedFekafnarFpgCMkgrtMyVIPFSmgpl
bb aaaaaaaaaaaaaaa bbbbbbbb
160 170 180 190 200
d1ii2a2 ( 102 ) e-ryrlkVrVfTTRPyHAlfMrdmlivPt-pelatFgepdYvIyNAGeck
d1j3ba2 ( 110 ) r-ryrlaVrVVTESPwHAlfArnmfilpr-rf-gaf-vpgFtVVHAPyfq
d2olra2 ( 129 ) p-dtrLsVRFITeVaWQAhfVknmFirPsdeelagF--pdFiVMNGAkct
d4gmua1 ( 150 ) gsplAkiGIELTDspyVVasMrimTrMGtsVleaLgdgeFIkCLHSVGcp
bbbbbbb aaaaaaaaaa bbbbbb
210 220 230 240 250
d1ii2a2 ( 151 ) Ad-p-----sipgLtstTCVALNFktreQVILgTeyagEMkkgiltVmfe
d1j3ba2 ( 162 ) Av-p-----erDgTrseVFVGISFqrrLVLIVGTkyagEIkkSIftVmny
d2olra2 ( 177 ) Npqw-----keqgLnsenFVAFNLterMQLIGGTwyGgEMkkGMfsMmny
d4gmua1 ( 200 ) lplkkplvnnWaCnpelTlIAHLPdrreIiSFGSGygGNSlLGkkCFalr
bbbbb bbbbb aaaaaaaaaaaaa
260
d1ii2a2 ( 195 ) lmpqmn
d1j3ba2 ( 206 ) lmpkrg
d2olra2 ( 222 ) llplkg
d4gmua1 ( 250 ) Iasrlakeeg
a
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Phosphoenolpyruvate (PEP) carboxykinase (ATP-oxaloacetate carboxy-lyase) | PEP carboxykinase N-terminal domain | Trypanosoma cruzi [TaxId: 5693] | 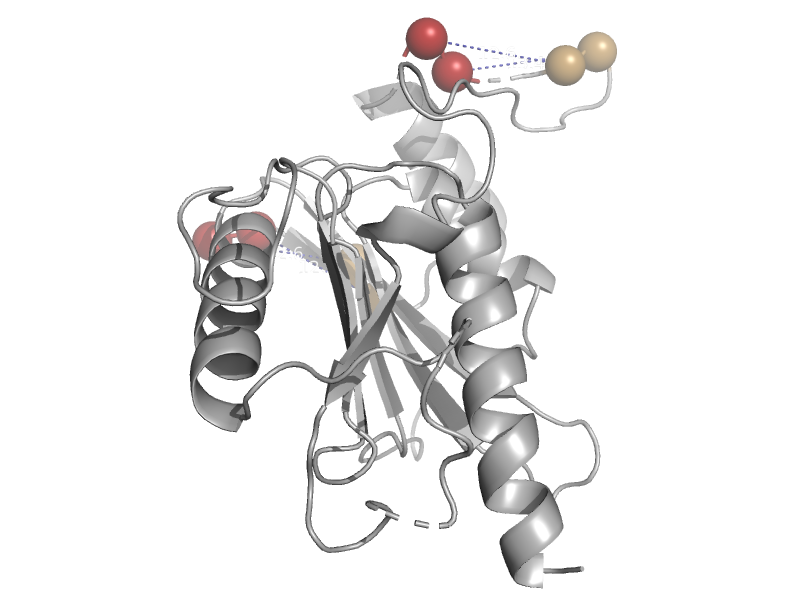 | view | |
| Phosphoenolpyruvate (PEP) carboxykinase (ATP-oxaloacetate carboxy-lyase) | PEP carboxykinase N-terminal domain | Thermus thermophilus [TaxId: 274] | 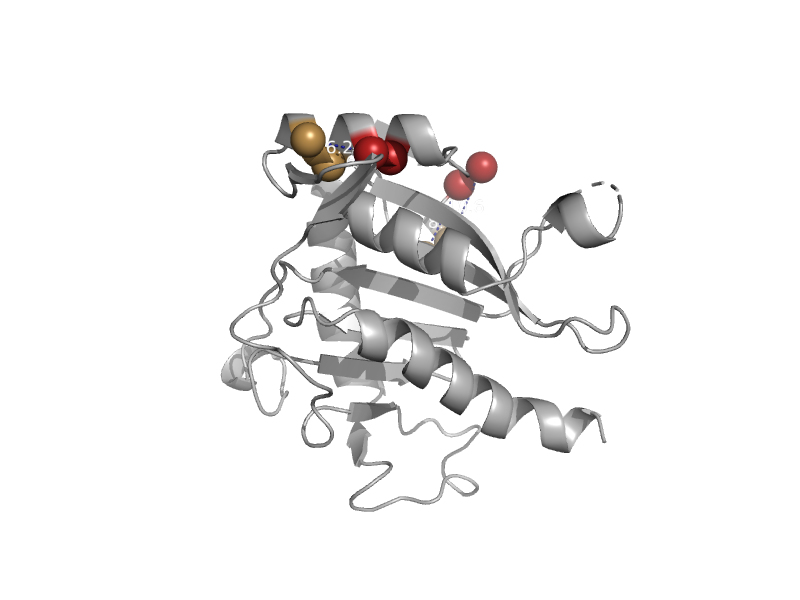 | view | |
| Phosphoenolpyruvate (PEP) carboxykinase (ATP-oxaloacetate carboxy-lyase) | PEP carboxykinase N-terminal domain | Escherichia coli [TaxId: 562] | 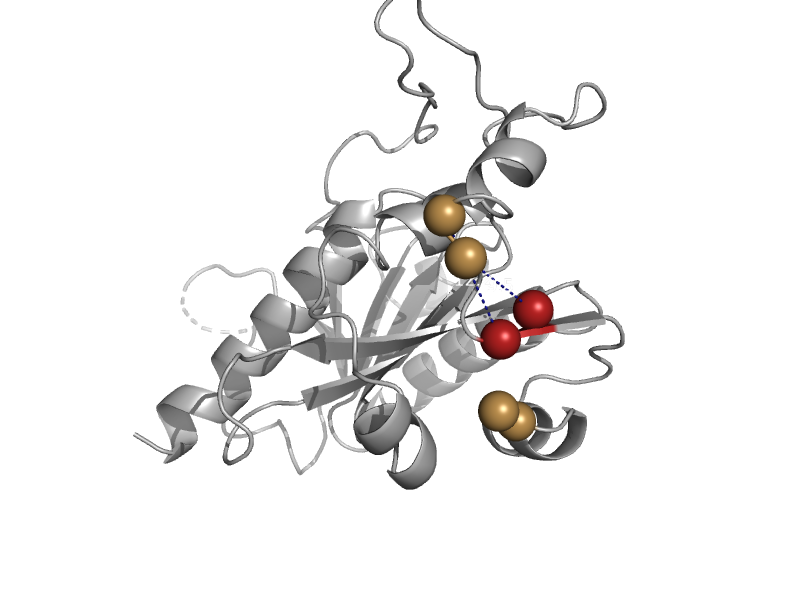 | view | |
| Cytosolic phosphoenolpyruvate carboxykinase (GTP-hydrolyzing) | PEP carboxykinase N-terminal domain | Norway rat (Rattus norvegicus) [TaxId: 10116] | 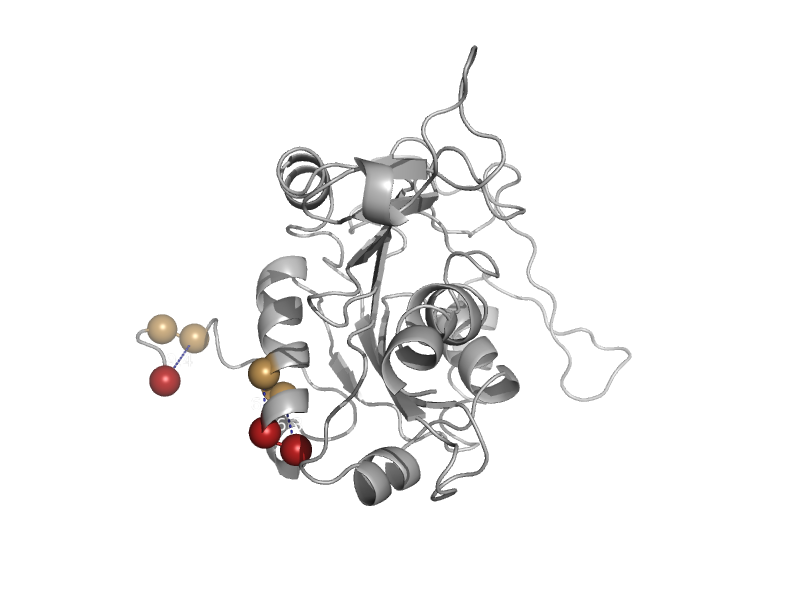 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | P | T | I | H | R | N | L | L | S | P | E | L | V | Q | W | A | L | K | - | - | - | - | - | I | E | D | S | - | L | T | A | R | - | - | G | A | L | A | V | M | S | Y | A | - | K | T | G | R | S | P | L | D | K | R | I | V | D | T | D | D | V | R | - | N | V | D | W | G | - | - | - | - | - | - | V | N | M | K | L | S | E | E | S | F | A | R | V | R | K | I | A | K | E | F | L | D | T | R | E | H | L | F | V | V | D | C | F | A | G | H | D | E | - | R | Y | R | L | K | V | R | V | F | T | T | R | P | Y | H | A | L | F | M | R | D | M | L | I | V | P | T | - | P | E | L | A | T | F | G | E | P | D | Y | V | I | Y | N | A | G | E | C | K | A | D | - | P | - | - | - | - | - | S | I | P | G | L | T | S | T | T | C | V | A | L | N | F | K | T | R | E | Q | V | I | L | G | T | E | Y | A | G | E | M | K | K | G | I | L | T | V | M | F | E | L | M | P | Q | M | N | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | - | - | - | - | R | L | E | A | L | G | - | - | I | H | P | K | K | R | V | F | W | N | T | V | S | P | V | L | V | E | H | T | L | L | - | - | - | - | - | R | G | E | G | L | L | A | H | H | - | - | G | P | L | V | V | D | T | T | P | - | Y | T | G | R | S | P | K | D | K | F | V | V | R | E | P | E | V | E | G | E | I | W | W | G | - | - | - | - | - | E | V | N | Q | P | F | A | P | E | A | F | E | A | L | Y | Q | R | V | V | Q | Y | L | S | E | R | - | D | L | Y | V | Q | D | L | Y | A | G | A | D | R | - | R | Y | R | L | A | V | R | V | V | T | E | S | P | W | H | A | L | F | A | R | N | M | F | I | L | P | R | - | R | F | - | G | A | F | - | V | P | G | F | T | V | V | H | A | P | Y | F | Q | A | V | - | P | - | - | - | - | - | E | R | D | G | T | R | S | E | V | F | V | G | I | S | F | Q | R | R | L | V | L | I | V | G | T | K | Y | A | G | E | I | K | K | S | I | F | T | V | M | N | Y | L | M | P | K | R | G | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | L | T | P | Q | E | L | E | A | Y | G | - | - | I | S | D | V | H | D | I | V | Y | N | P | S | Y | D | L | L | Y | Q | E | E | L | D | P | S | L | T | G | Y | E | R | G | V | L | T | N | L | - | - | G | A | V | A | V | D | T | G | I | - | F | T | G | R | S | P | K | D | K | Y | I | V | R | D | D | T | T | R | D | T | F | W | W | A | D | - | G | G | K | N | D | N | K | P | L | S | P | E | T | W | Q | H | L | K | G | L | V | T | R | Q | L | S | G | K | - | R | L | F | V | V | D | A | F | C | G | A | N | P | - | D | T | R | L | S | V | R | F | I | T | E | V | A | W | Q | A | H | F | V | K | N | M | F | I | R | P | S | D | E | E | L | A | G | F | - | - | P | D | F | I | V | M | N | G | A | K | C | T | N | P | Q | W | - | - | - | - | - | K | E | Q | G | L | N | S | E | N | F | V | A | F | N | L | T | E | R | M | Q | L | I | G | G | T | W | Y | G | G | E | M | K | K | G | M | F | S | M | M | N | Y | L | L | P | L | K | G | - | - | - | - |
| P | Q | L | H | N | G | L | D | F | S | A | K | V | I | Q | G | S | L | D | S | L | P | Q | E | V | R | K | F | V | E | G | N | A | Q | L | C | Q | P | E | Y | I | H | I | C | D | G | S | E | E | E | Y | G | R | L | L | A | H | M | Q | E | - | - | E | G | V | I | R | K | L | K | K | Y | D | N | C | W | L | A | L | T | D | P | R | D | V | A | R | I | E | S | K | T | V | I | I | - | T | Q | E | Q | R | D | T | V | P | I | P | K | S | G | Q | S | Q | L | G | R | W | M | S | E | E | D | F | E | K | A | F | N | A | R | F | P | G | C | M | K | G | R | T | M | Y | V | I | P | F | S | M | G | P | L | G | S | P | L | A | K | I | G | I | E | L | T | D | S | P | Y | V | V | A | S | M | R | I | M | T | R | M | G | T | S | V | L | E | A | L | G | D | G | E | F | I | K | C | L | H | S | V | G | C | P | L | P | L | K | K | P | L | V | N | N | W | A | C | N | P | E | L | T | L | I | A | H | L | P | D | R | R | E | I | I | S | F | G | S | G | Y | G | G | N | S | L | L | G | K | K | C | F | A | L | R | I | A | S | R | L | A | K | E | E | G |
