solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1uz5a1 ( 329 ) igkkvA-lk-hvfs---rrqflpVkleg------dlAvpilk-gsgavt
d1wu2a1 ( 325 ) evkvkAiLqddIpSqlgryefikIyyen------giArvikkkgsgils
d2nqra1 ( 327 ) lparqrVrTasrLkKtpgrldfQrGvlqrnad-geleVttTghqgshifs
d6fgca3 ( 654 ) ptiikArlscdvkldp-rpeyhrCiltwhhqeplPwAqstg------lm
bbbbbb bbbbbbbb bbbb
60 70 80
d1uz5a1 ( 371 ) sfidadGfveipet-------degeeveVtlfkg---w
d1wu2a1 ( 368 ) SllaSnAyLeIpe---dsegyrrgeeVwITly
d2nqra1 ( 376 ) SfslGnCfIVLer---drgnVevgewVeVepFnalfgg
d6fgca3 ( 702 ) smrsanGllmlppkteqyvelhkgevvdvmvigrl
aaaa bbbbb bbbbbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Molybdenum cofactor biosynthesis protein MoeA, C-terminal domain | MoeA C-terminal domain-like | Pyrococcus horikoshii, PH0582 [TaxId: 53953] | 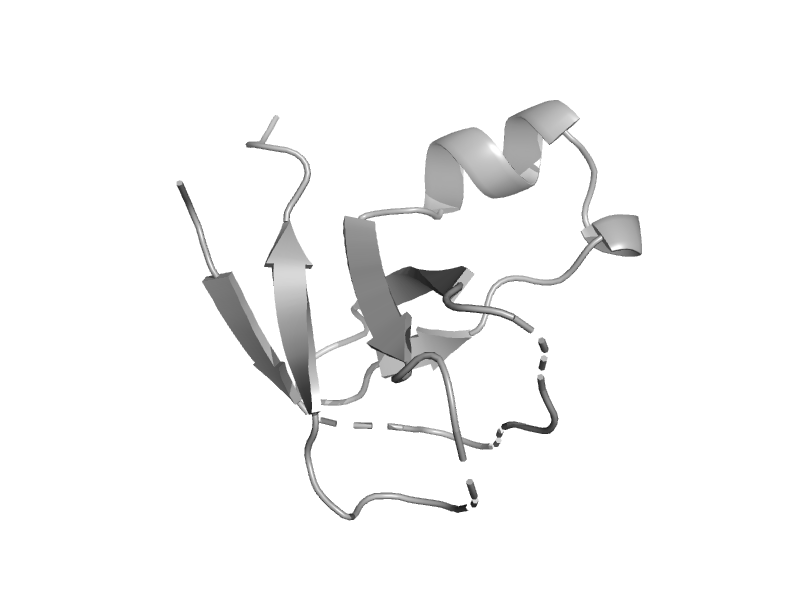 | view | |
| Molybdenum cofactor biosynthesis protein MoeA, C-terminal domain | MoeA C-terminal domain-like | Pyrococcus horikoshii, PH1647 [TaxId: 53953] | 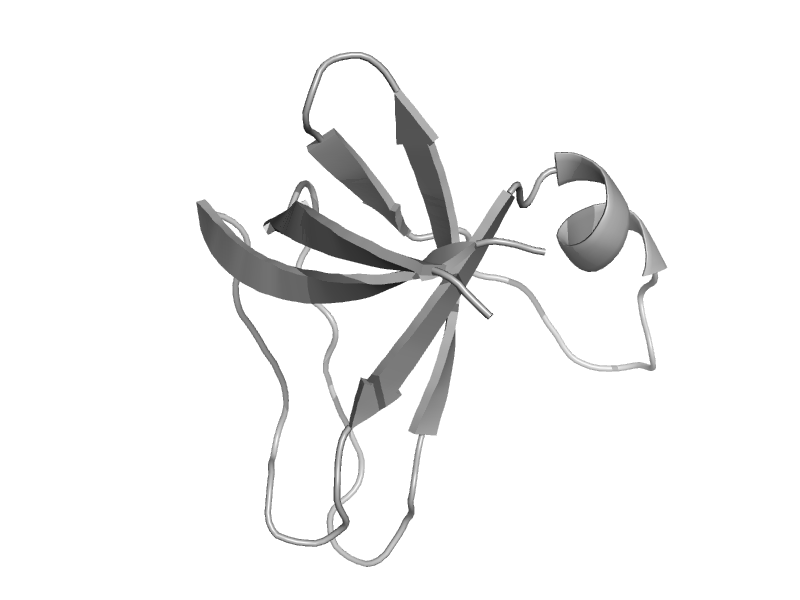 | view | |
| Molybdenum cofactor biosynthesis protein MoeA, C-terminal domain | MoeA C-terminal domain-like | Escherichia coli [TaxId: 562] | 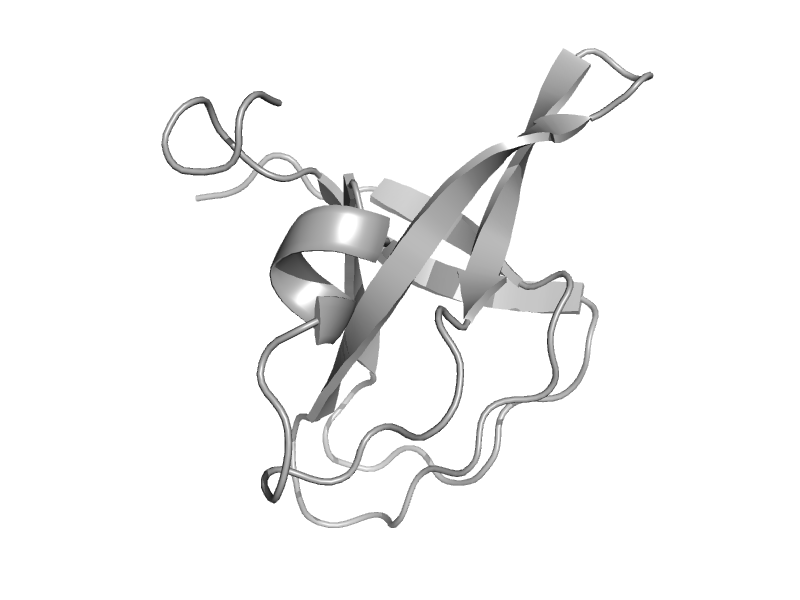 | view | |
| automated matches | automated matches | Norway rat (Rattus norvegicus) [TaxId: 10116] | 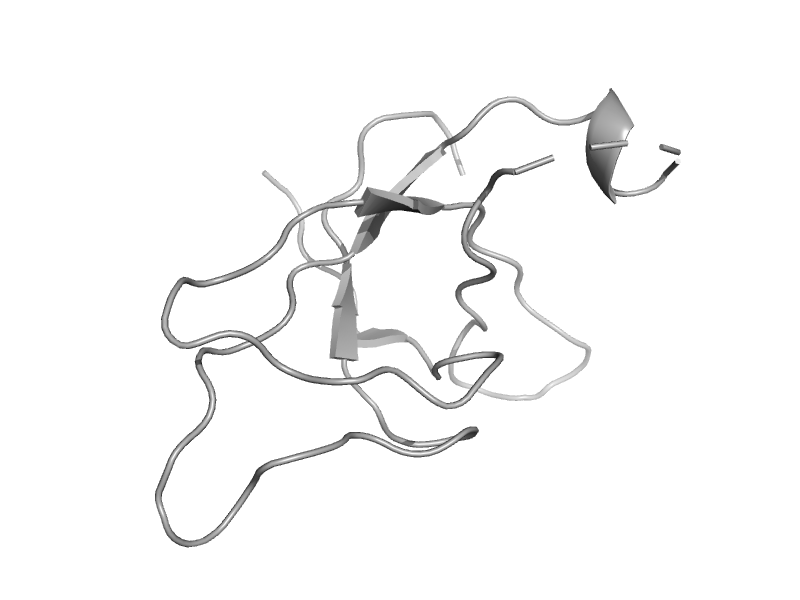 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | I | G | K | K | V | A | - | L | K | - | H | V | F | S | - | - | - | R | R | Q | F | L | P | V | K | L | E | G | - | - | - | - | - | - | D | L | A | V | P | I | L | K | - | G | S | G | A | V | T | S | F | I | D | A | D | G | F | V | E | I | P | E | T | - | - | - | - | - | - | - | D | E | G | E | E | V | E | V | T | L | F | K | G | - | - | - | W |
| - | E | V | K | V | K | A | I | L | Q | D | D | I | P | S | Q | L | G | R | Y | E | F | I | K | I | Y | Y | E | N | - | - | - | - | - | - | G | I | A | R | V | I | K | K | K | G | S | G | I | L | S | S | L | L | A | S | N | A | Y | L | E | I | P | E | - | - | - | D | S | E | G | Y | R | R | G | E | E | V | W | I | T | L | Y | - | - | - | - | - | - |
| L | P | A | R | Q | R | V | R | T | A | S | R | L | K | K | T | P | G | R | L | D | F | Q | R | G | V | L | Q | R | N | A | D | - | G | E | L | E | V | T | T | T | G | H | Q | G | S | H | I | F | S | S | F | S | L | G | N | C | F | I | V | L | E | R | - | - | - | D | R | G | N | V | E | V | G | E | W | V | E | V | E | P | F | N | A | L | F | G | G |
| - | P | T | I | I | K | A | R | L | S | C | D | V | K | L | D | P | - | R | P | E | Y | H | R | C | I | L | T | W | H | H | Q | E | P | L | P | W | A | Q | S | T | G | - | - | - | - | - | - | L | M | S | M | R | S | A | N | G | L | L | M | L | P | P | K | T | E | Q | Y | V | E | L | H | K | G | E | V | V | D | V | M | V | I | G | R | L | - | - | - |
