solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1lsha2 ( 18 ) FqpgkVYrYsYdafsIsglpepgvnraglsgeMkIeIhG
d1lsha3 ( 621 ) cryskavhvdtfnartmagvsadyfriNS-psgplpravaakirgqgm--
bbbbbbbbbbbb bbbbbbbbbbbb
60 70 80 90 100
d1lsha2 ( 57 ) hthnqAtLkItqVnlkyF-----lgpwpsdsfypltggydhfiqqLevpV
d1lsha3 ( 668 ) --g---yasdivefGlraeglQellyd--wksvpe---------erplas
bbbbbbbbbbb b
110 120 130 140 150
d1lsha2 ( 102 ) RFdysagrIgdIyAppqVtdtAVNiVRgILNLFQLsLkknqqtfelqEtG
d1lsha3 ( 741 ) gyvkvhgqevvfaeldkkmqeqigavVskle----------qgmdvlltk
bbbbb bbbbbb aaaaaaaaaaa bbbbbbb
160 170 180 190 200
d1lsha2 ( 152 ) veGiCqTtYvvqegyr---tnemaVvKtKdLnnCdhkvy-ktmgtayae-
d1lsha3 ( 799 ) ---gyvvsevrymqpvcigipmDlnllvsGvttnranlsAsFs------s
bbbbbbbbbb bbbbbbbbbbbbbbbb
210 220 230 240 250
d1lsha2 ( 197 ) ----rçptçqkm-----nkn---lrStAvYnYaIfdepsgyiIksAhSee
d1lsha3 ( 841 ) lpadmkladllatnielrvaattsmsqhavaimglttd-lakagmqthyk
aaaa bbbbbbbbbbbbb bbbbbbbbb
260 270 280 290 300
d1lsha2 ( 235 ) iqqlSv------fdike-------------gnvviesrqkLiLeg----i
d1lsha3 ( 890 ) tsaglgvngkiemnaresnfkaslkpfqqktvvvlstMeSivFvrdpsgs
bbbb bbbbbbbbbbbbb b
310 320 330 340 350
d1lsha2 ( 262 ) qsap----------------------------------------------
d1lsha3 ( 940 ) rilpvLppkmtqkqihdimtarPvmrrkqscsksaalsskvCfsarlrNA
bb
360 370 380 390
d1lsha2 ( 266 ) -----------aasqaAsl-------qnrggLmykfp
d1lsha3 (1034 ) afirnallykitGdyvskvyvqptsskaqitkvelelqag
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Lipovitellin-phosvitin complex; beta-sheet shell regions | Lipovitellin-phosvitin complex; beta-sheet shell regions | Lamprey (Ichthyomyzon unicuspis) [TaxId: 30308] | 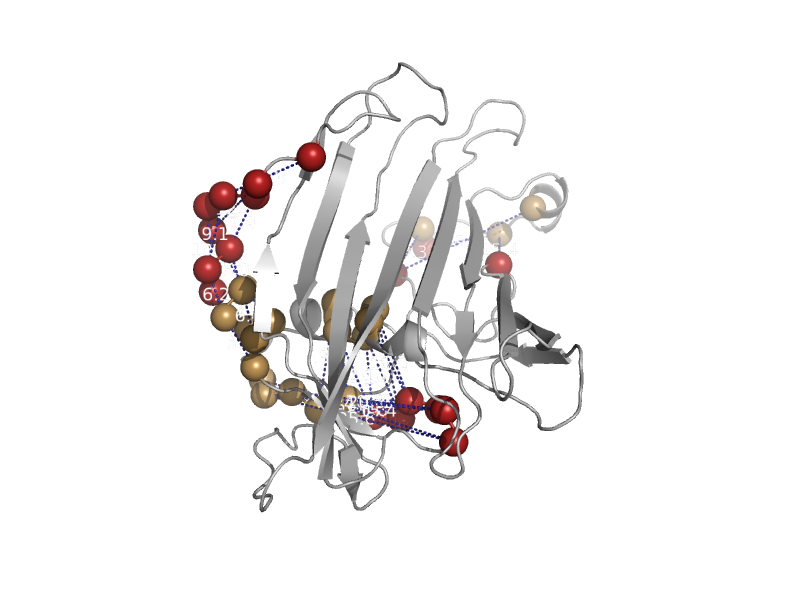 | view | |
| Lipovitellin-phosvitin complex; beta-sheet shell regions | Lipovitellin-phosvitin complex; beta-sheet shell regions | Lamprey (Ichthyomyzon unicuspis) [TaxId: 30308] | 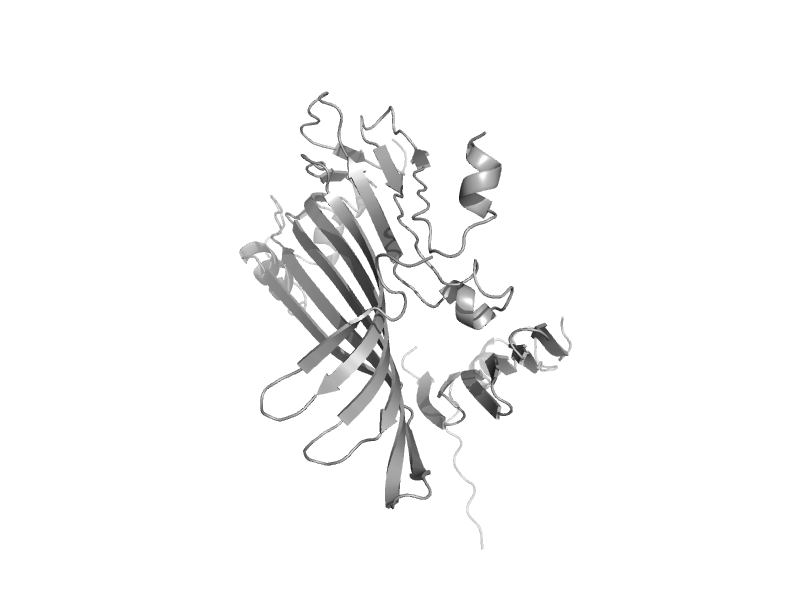 | view |
| Domain ID | Name |
|---|---|
| d1lshb- | 1lsh B: |
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | 330 | 340 | 350 | 360 | 370 | 380 | 390 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | F | Q | P | G | K | V | Y | R | Y | S | Y | D | A | F | S | I | S | G | L | P | E | P | G | V | N | R | A | G | L | S | G | E | M | K | I | E | I | H | G | H | T | H | N | Q | A | T | L | K | I | T | Q | V | N | L | K | Y | F | - | - | - | - | - | L | G | P | W | P | S | D | S | F | Y | P | L | T | G | G | Y | D | H | F | I | Q | Q | L | E | V | P | V | R | F | D | Y | S | A | G | R | I | G | D | I | Y | A | P | P | Q | V | T | D | T | A | V | N | I | V | R | G | I | L | N | L | F | Q | L | S | L | K | K | N | Q | Q | T | F | E | L | Q | E | T | G | V | E | G | I | C | Q | T | T | Y | V | V | Q | E | G | Y | R | - | - | - | T | N | E | M | A | V | V | K | T | K | D | L | N | N | C | D | H | K | V | Y | - | K | T | M | G | T | A | Y | A | E | - | - | - | - | - | R | C | P | T | C | Q | K | M | - | - | - | - | - | N | K | N | - | - | - | L | R | S | T | A | V | Y | N | Y | A | I | F | D | E | P | S | G | Y | I | I | K | S | A | H | S | E | E | I | Q | Q | L | S | V | - | - | - | - | - | - | F | D | I | K | E | - | - | - | - | - | - | - | - | - | - | - | - | - | G | N | V | V | I | E | S | R | Q | K | L | I | L | E | G | - | - | - | - | I | Q | S | A | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | A | S | Q | A | A | S | L | - | - | - | - | - | - | - | Q | N | R | G | G | L | M | Y | K | F | P | - | - | - |
| C | R | Y | S | K | A | V | H | V | D | T | F | N | A | R | T | M | A | G | V | S | A | D | Y | F | R | I | N | S | - | P | S | G | P | L | P | R | A | V | A | A | K | I | R | G | Q | G | M | - | - | - | - | G | - | - | - | Y | A | S | D | I | V | E | F | G | L | R | A | E | G | L | Q | E | L | L | Y | D | - | - | W | K | S | V | P | E | - | - | - | - | - | - | - | - | - | E | R | P | L | A | S | G | Y | V | K | V | H | G | Q | E | V | V | F | A | E | L | D | K | K | M | Q | E | Q | I | G | A | V | V | S | K | L | E | - | - | - | - | - | - | - | - | - | - | Q | G | M | D | V | L | L | T | K | - | - | - | G | Y | V | V | S | E | V | R | Y | M | Q | P | V | C | I | G | I | P | M | D | L | N | L | L | V | S | G | V | T | T | N | R | A | N | L | S | A | S | F | S | - | - | - | - | - | - | S | L | P | A | D | M | K | L | A | D | L | L | A | T | N | I | E | L | R | V | A | A | T | T | S | M | S | Q | H | A | V | A | I | M | G | L | T | T | D | - | L | A | K | A | G | M | Q | T | H | Y | K | T | S | A | G | L | G | V | N | G | K | I | E | M | N | A | R | E | S | N | F | K | A | S | L | K | P | F | Q | Q | K | T | V | V | V | L | S | T | M | E | S | I | V | F | V | R | D | P | S | G | S | R | I | L | P | V | L | P | P | K | M | T | Q | K | Q | I | H | D | I | M | T | A | R | P | V | M | R | R | K | Q | S | C | S | K | S | A | A | L | S | S | K | V | C | F | S | A | R | L | R | N | A | A | F | I | R | N | A | L | L | Y | K | I | T | G | D | Y | V | S | K | V | Y | V | Q | P | T | S | S | K | A | Q | I | T | K | V | E | L | E | L | Q | A | G |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1lsha2 | A | Q91062 | GO:0005319 |
| d1lsha2 | A | Q91062 | GO:0006869 |
| d1lsha2 | A | Q91062 | GO:0005319 |
| d1lsha2 | A | Q91062 | GO:0006869 |
| d1lsha2 | A | Q91062 | GO:0005319 |
| d1lsha2 | A | Q91062 | GO:0006869 |
| d1lsha2 | A | Q91062 | GO:0005319 |
| d1lsha2 | A | Q91062 | GO:0006869 |
| d1lsha3 | A | Q91062 | GO:0005319 |
| d1lsha3 | A | Q91062 | GO:0006869 |
| d1lsha3 | A | Q91062 | GO:0005319 |
| d1lsha3 | A | Q91062 | GO:0006869 |
| d1lsha3 | A | Q91062 | GO:0005319 |
| d1lsha3 | A | Q91062 | GO:0006869 |
| d1lsha3 | A | Q91062 | GO:0005319 |
| d1lsha3 | A | Q91062 | GO:0006869 |
