solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1gnla- ( 1 ) snamfcYQcQeTvgnkGctqvGvc--------gkkpeTAaLqdALIyV--
d2yivx- ( 2 ) arqnlk--------StDrAvqqMldkAkregi
d3i01m- ( 2 ) tdFdkiFegaipegkePvaLFreVyhGAit--
aaaaaaaaaaaaa
60 70 80 90 100
d1gnla- ( 41 ) --TKGLGqIA----trLraegk---------------------------a
d2yivx- ( 26 ) qtvWdryeamkpqcgfgetglccrhclqGpcrinpfgdepkvGic---ga
d3i01m- ( 32 ) --aTsyAeil----Lnqairtygp---dhpVgYpdTayyLPVIrCFsgee
aaaaaaaa aaaaa
110 120 130 140 150
d1gnla- ( 58 ) VdhriDrlVTGNLFATIt-----NANfdddiLAerVrmTCaaKkelaasL
d2yivx- ( 73 ) taevivARglDrsiAaGA-----ag--HSgHAkhlAhtlkkAvqg-----
d3i01m- ( 73 ) VkkLgdLppILnrkraqVspvlnfenARlAGEATWYAaEIIEALrYlkyk
aaaaaaaaa aaaaaaaaaaaaaaa
160 170 180 190 200
d1gnla- ( 103 ) tdksgLsdAAlwease-ksaMlakAgtVgVmatt--------dddvrSLr
d2yivx- ( 111 ) -------kAasYmikd-rtklhsiAk---rlgiptegq----kdediAle
d3i01m- ( 123 ) pdepllpppwTgFIgDpvVRrFGikMVdwtIPGEAiILgrAkdskaLakI
aaaa aaa aaaaaaa
210 220 230 240 250
d1gnla- ( 144 ) wLITFGLKG-----MAAYAKhAdvLgkhe---------------nsLdaF
d2yivx- ( 146 ) vAkaAlaDF-----hekdtpvlWvttvlppsRvkvlsa-hgliPaGidhE
d3i01m- ( 173 ) VkeLMGmGFMLFICdeAVeQLlee-----nvkLgidyIAYPLGnfTqiVH
aaaaaa aa aa
260 270 280 290 300
d1gnla- ( 174 ) MQeALakTld--dsl-svadLvaLtleTGkfG--------vsAMalLdaA
d2yivx- ( 190 ) iaeimhrtsm--gcdadaqnlllGGlrCSlAD--------lAGCyMGtDl
d3i01m- ( 218 ) AANYaLRAGMMFGgvtpgareeQRdYQrrrIrAFVLYLgehdmvkTAaAF
aaaaaaa aaaaaaaaaaa aaaaaaaa
310 320 330 340 350
d1gnla- ( 213 ) NtgtYghPeitkVn----IgvgsnpGILISGhdLrDLeMLLkqTegt---
d2yivx- ( 230 ) ADilFgtPapvvteSnlGvlkadAvnvAvHGHnPvlSDiivsvSkeMene
d3i01m- ( 268 ) GAIFTGFPVITDqpLpedkqIpdWFfsve------dydkIVqiAmetRgI
aaaa b bb aaaaa
360 370 380 390 400
d1gnla- ( 256 ) -------gVDVYTHSEMLPAhYYpaFkkyahFkGNYGnAWwkQkeeFesF
d2yivx- ( 280 ) AraagAtginvvGiCCtGnEvlMRhg-------ipACtHSvSqEMAMitg
d3i01m- ( 312 ) kltkikldLpInfgPAFegEsIrkg-------dMyVEMGg---------n
410 420 430 440 450
d1gnla- ( 299 ) nG-PVLLTTNcLVpPk----dsYkdrVYTTg------iVgftgCkhipge
d2yivx- ( 323 ) AlDAMilDYqciqpsvAtiaectgttvittme-----mskitgAthvnFa
d3i01m- ( 346 ) rtpAFELVrtvs----eseItdgkievigpdIdqipegsklpLGILVdIy
bbb bb bb
460 470 480 490 500
d1gnla- ( 338 ) iGehK-dFsaIiahAktc-------paPteiesgeiiGGFAhnqvlalad
d2yivx- ( 368 ) eeaAvenAkqilrlAidtfkrrkgkpveiPniktkvvAGFSteaiinAls
d3i01m- ( 392 ) GrkMqadFEgVLErrIhdFINyGegLwHtgqrn-iNwLrVskdAvakgFr
aaaaaa a bbbb aaaaaa
510 520 530 540 550
d1gnla- ( 380 ) ---------kVidavksgaIkkFVvMAgcDGra-k-sRsYYtdFAegLpk
d2yivx- ( 418 ) klnandplkPlidnvvngnirGvClFagcnnvkvp-qDqnFttiArkllk
d3i01m- ( 441 ) FknYGeILvakMkeefpaIVdrVqVtIFTdeakVkeymevArekYkerdd
aaaaaa bbb aaaaaaaa
560 570 580 590 600
d1gnla- ( 419 ) ----------dTVIlTAGCAKYrYnklnLg----------dig-------
d2yivx- ( 467 ) ---------qnvlvvAtGcGAGAlMRhGFMdpanvdel--Cgdglkavlt
d3i01m- ( 491 ) rmrgltDetvdtFYScvlcqsFApnHVcIVTPeRvgLcgavswldAkAsy
bbbb
610 620 630 640 650
d1gnla- ( 442 ) ---------------------------------gipRvLDAGQCNDSySL
d2yivx- ( 506 ) aiGeangl-----------------------ggplPPvlHMGScvDnSRA
d3i01m- ( 541 ) einhagpNqpIpkegeidpikGIWksVndyLytaSnrnleqVCLYTLmen
660 670 680 690 700
d1gnla- ( 459 ) avIalkLkevfgle-dvndLpIvyNIA-W--YEQKAVIvlLAlLslgVkn
d2yivx- ( 533 ) valvaalAnrlgv--dldrlPvvASaAEA--mheKavAiGtWAvtiGl-P
d3i01m- ( 591 ) PMTSCGCFe-AIMAiLpeCnGIMITtrdhagmTPSgmtFStLAgmIGGgt
333 bbbbb aaaaaaa
710 720 730 740 750
d1gnla- ( 505 ) IhLGPtlPaFlspnVakvlve----qfn--IggitspqdDlkaff
d2yivx- ( 578 ) tHiGvlppitgSlpvtqiltssvkditgGyFiveldpetAAdkllaAine
d3i01m- ( 640 ) qtpGfMGIGrt-YIvskkFIsadggIaRIVWMP----ksLkdfLhdeFvr
aaaaaa
760 770 780 790
d1gnla-
d2yivx- ( 628 ) rRaglglpw
d3i01m- ( 685 ) RSveeglgedFidkIADetigttvdeIlpyLeekgHpAltmdpIm
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Hybrid cluster protein (prismane protein) | Hybrid cluster protein (prismane protein) | Desulfovibrio desulfuricans [TaxId: 876] | 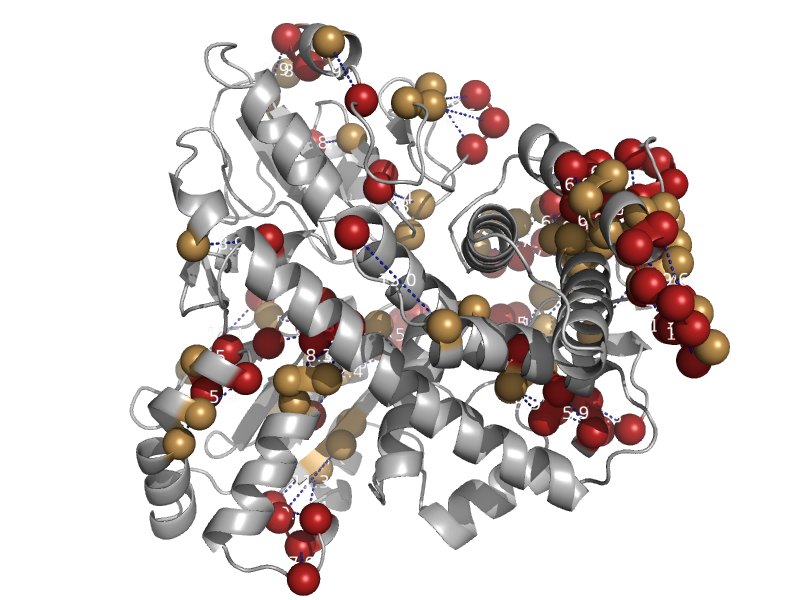 | view | |
| Ni-containing carbon monoxide dehydrogenase | Carbon monoxide dehydrogenase | Carboxydothermus hydrogenoformans [TaxId: 129958] | 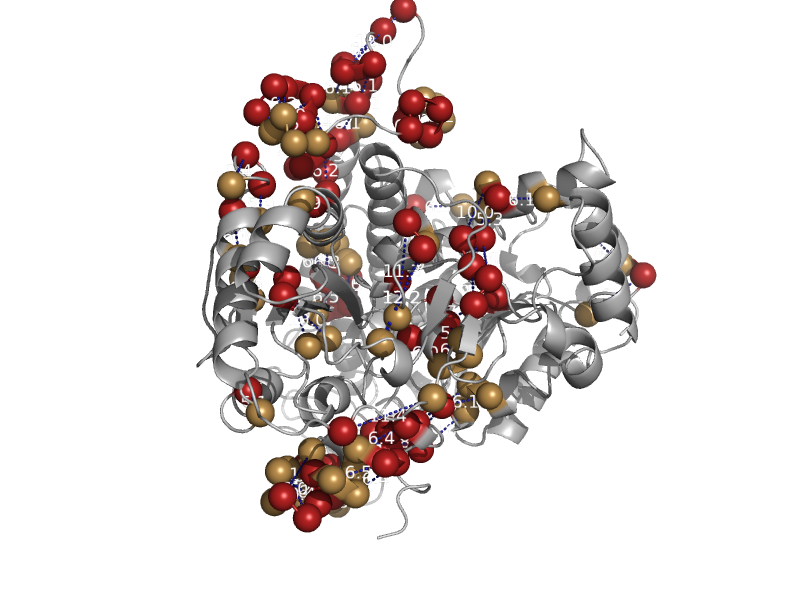 | view | |
| automated matches | Acetyl-CoA synthase | Moorella thermoacetica [TaxId: 1525] | 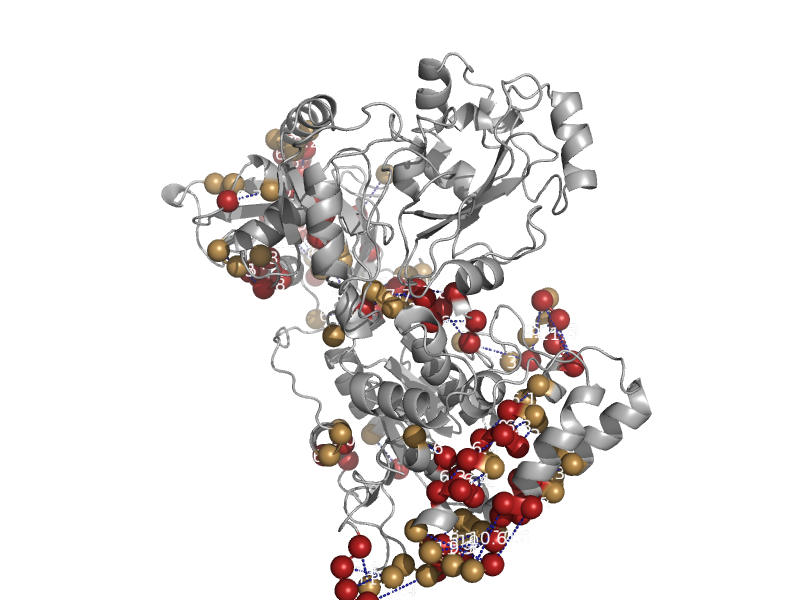 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | 330 | 340 | 350 | 360 | 370 | 380 | 390 | 400 | 410 | 420 | 430 | 440 | 450 | 460 | 470 | 480 | 490 | 500 | 510 | 520 | 530 | 540 | 550 | 560 | 570 | 580 | 590 | 600 | 610 | 620 | 630 | 640 | 650 | 660 | 670 | 680 | 690 | 700 | 710 | 720 | 730 | 740 | 750 | 760 | 770 | 780 | 790 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | N | A | M | F | C | Y | Q | C | Q | E | T | V | G | N | K | G | C | T | Q | V | G | V | C | - | - | - | - | - | - | - | - | G | K | K | P | E | T | A | A | L | Q | D | A | L | I | Y | V | - | - | - | - | T | K | G | L | G | Q | I | A | - | - | - | - | T | R | L | R | A | E | G | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | V | D | H | R | I | D | R | L | V | T | G | N | L | F | A | T | I | T | - | - | - | - | - | N | A | N | F | D | D | D | I | L | A | E | R | V | R | M | T | C | A | A | K | K | E | L | A | A | S | L | T | D | K | S | G | L | S | D | A | A | L | W | E | A | S | E | - | K | S | A | M | L | A | K | A | G | T | V | G | V | M | A | T | T | - | - | - | - | - | - | - | - | D | D | D | V | R | S | L | R | W | L | I | T | F | G | L | K | G | - | - | - | - | - | M | A | A | Y | A | K | H | A | D | V | L | G | K | H | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | S | L | D | A | F | M | Q | E | A | L | A | K | T | L | D | - | - | D | S | L | - | S | V | A | D | L | V | A | L | T | L | E | T | G | K | F | G | - | - | - | - | - | - | - | - | V | S | A | M | A | L | L | D | A | A | N | T | G | T | Y | G | H | P | E | I | T | K | V | N | - | - | - | - | I | G | V | G | S | N | P | G | I | L | I | S | G | H | D | L | R | D | L | E | M | L | L | K | Q | T | E | G | T | - | - | - | - | - | - | - | - | - | - | G | V | D | V | Y | T | H | S | E | M | L | P | A | H | Y | Y | P | A | F | K | K | Y | A | H | F | K | G | N | Y | G | N | A | W | W | K | Q | K | E | E | F | E | S | F | N | G | - | P | V | L | L | T | T | N | C | L | V | P | P | K | - | - | - | - | D | S | Y | K | D | R | V | Y | T | T | G | - | - | - | - | - | - | I | V | G | F | T | G | C | K | H | I | P | G | E | I | G | E | H | K | - | D | F | S | A | I | I | A | H | A | K | T | C | - | - | - | - | - | - | - | P | A | P | T | E | I | E | S | G | E | I | I | G | G | F | A | H | N | Q | V | L | A | L | A | D | - | - | - | - | - | - | - | - | - | K | V | I | D | A | V | K | S | G | A | I | K | K | F | V | V | M | A | G | C | D | G | R | A | - | K | - | S | R | S | Y | Y | T | D | F | A | E | G | L | P | K | - | - | - | - | - | - | - | - | - | - | D | T | V | I | L | T | A | G | C | A | K | Y | R | Y | N | K | L | N | L | G | - | - | - | - | - | - | - | - | - | - | D | I | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | I | P | R | V | L | D | A | G | Q | C | N | D | S | Y | S | L | A | V | I | A | L | K | L | K | E | V | F | G | L | E | - | D | V | N | D | L | P | I | V | Y | N | I | A | - | W | - | - | Y | E | Q | K | A | V | I | V | L | L | A | L | L | S | L | G | V | K | N | I | H | L | G | P | T | L | P | A | F | L | S | P | N | V | A | K | V | L | V | E | - | - | - | - | Q | F | N | - | - | I | G | G | I | T | S | P | Q | D | D | L | K | A | F | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | R | Q | N | L | K | - | - | - | - | - | - | - | - | S | T | D | R | A | V | Q | Q | M | L | D | K | A | K | R | E | G | I | Q | T | V | W | D | R | Y | E | A | M | K | P | Q | C | G | F | G | E | T | G | L | C | C | R | H | C | L | Q | G | P | C | R | I | N | P | F | G | D | E | P | K | V | G | I | C | - | - | - | G | A | T | A | E | V | I | V | A | R | G | L | D | R | S | I | A | A | G | A | - | - | - | - | - | A | G | - | - | H | S | G | H | A | K | H | L | A | H | T | L | K | K | A | V | Q | G | - | - | - | - | - | - | - | - | - | - | - | - | K | A | A | S | Y | M | I | K | D | - | R | T | K | L | H | S | I | A | K | - | - | - | R | L | G | I | P | T | E | G | Q | - | - | - | - | K | D | E | D | I | A | L | E | V | A | K | A | A | L | A | D | F | - | - | - | - | - | H | E | K | D | T | P | V | L | W | V | T | T | V | L | P | P | S | R | V | K | V | L | S | A | - | H | G | L | I | P | A | G | I | D | H | E | I | A | E | I | M | H | R | T | S | M | - | - | G | C | D | A | D | A | Q | N | L | L | L | G | G | L | R | C | S | L | A | D | - | - | - | - | - | - | - | - | L | A | G | C | Y | M | G | T | D | L | A | D | I | L | F | G | T | P | A | P | V | V | T | E | S | N | L | G | V | L | K | A | D | A | V | N | V | A | V | H | G | H | N | P | V | L | S | D | I | I | V | S | V | S | K | E | M | E | N | E | A | R | A | A | G | A | T | G | I | N | V | V | G | I | C | C | T | G | N | E | V | L | M | R | H | G | - | - | - | - | - | - | - | I | P | A | C | T | H | S | V | S | Q | E | M | A | M | I | T | G | A | L | D | A | M | I | L | D | Y | Q | C | I | Q | P | S | V | A | T | I | A | E | C | T | G | T | T | V | I | T | T | M | E | - | - | - | - | - | M | S | K | I | T | G | A | T | H | V | N | F | A | E | E | A | A | V | E | N | A | K | Q | I | L | R | L | A | I | D | T | F | K | R | R | K | G | K | P | V | E | I | P | N | I | K | T | K | V | V | A | G | F | S | T | E | A | I | I | N | A | L | S | K | L | N | A | N | D | P | L | K | P | L | I | D | N | V | V | N | G | N | I | R | G | V | C | L | F | A | G | C | N | N | V | K | V | P | - | Q | D | Q | N | F | T | T | I | A | R | K | L | L | K | - | - | - | - | - | - | - | - | - | Q | N | V | L | V | V | A | T | G | C | G | A | G | A | L | M | R | H | G | F | M | D | P | A | N | V | D | E | L | - | - | C | G | D | G | L | K | A | V | L | T | A | I | G | E | A | N | G | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | G | P | L | P | P | V | L | H | M | G | S | C | V | D | N | S | R | A | V | A | L | V | A | A | L | A | N | R | L | G | V | - | - | D | L | D | R | L | P | V | V | A | S | A | A | E | A | - | - | M | H | E | K | A | V | A | I | G | T | W | A | V | T | I | G | L | - | P | T | H | I | G | V | L | P | P | I | T | G | S | L | P | V | T | Q | I | L | T | S | S | V | K | D | I | T | G | G | Y | F | I | V | E | L | D | P | E | T | A | A | D | K | L | L | A | A | I | N | E | R | R | A | G | L | G | L | P | W | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | D | F | D | K | I | F | E | G | A | I | P | E | G | K | E | P | V | A | L | F | R | E | V | Y | H | G | A | I | T | - | - | - | - | A | T | S | Y | A | E | I | L | - | - | - | - | L | N | Q | A | I | R | T | Y | G | P | - | - | - | D | H | P | V | G | Y | P | D | T | A | Y | Y | L | P | V | I | R | C | F | S | G | E | E | V | K | K | L | G | D | L | P | P | I | L | N | R | K | R | A | Q | V | S | P | V | L | N | F | E | N | A | R | L | A | G | E | A | T | W | Y | A | A | E | I | I | E | A | L | R | Y | L | K | Y | K | P | D | E | P | L | L | P | P | P | W | T | G | F | I | G | D | P | V | V | R | R | F | G | I | K | M | V | D | W | T | I | P | G | E | A | I | I | L | G | R | A | K | D | S | K | A | L | A | K | I | V | K | E | L | M | G | M | G | F | M | L | F | I | C | D | E | A | V | E | Q | L | L | E | E | - | - | - | - | - | N | V | K | L | G | I | D | Y | I | A | Y | P | L | G | N | F | T | Q | I | V | H | A | A | N | Y | A | L | R | A | G | M | M | F | G | G | V | T | P | G | A | R | E | E | Q | R | D | Y | Q | R | R | R | I | R | A | F | V | L | Y | L | G | E | H | D | M | V | K | T | A | A | A | F | G | A | I | F | T | G | F | P | V | I | T | D | Q | P | L | P | E | D | K | Q | I | P | D | W | F | F | S | V | E | - | - | - | - | - | - | D | Y | D | K | I | V | Q | I | A | M | E | T | R | G | I | K | L | T | K | I | K | L | D | L | P | I | N | F | G | P | A | F | E | G | E | S | I | R | K | G | - | - | - | - | - | - | - | D | M | Y | V | E | M | G | G | - | - | - | - | - | - | - | - | - | N | R | T | P | A | F | E | L | V | R | T | V | S | - | - | - | - | E | S | E | I | T | D | G | K | I | E | V | I | G | P | D | I | D | Q | I | P | E | G | S | K | L | P | L | G | I | L | V | D | I | Y | G | R | K | M | Q | A | D | F | E | G | V | L | E | R | R | I | H | D | F | I | N | Y | G | E | G | L | W | H | T | G | Q | R | N | - | I | N | W | L | R | V | S | K | D | A | V | A | K | G | F | R | F | K | N | Y | G | E | I | L | V | A | K | M | K | E | E | F | P | A | I | V | D | R | V | Q | V | T | I | F | T | D | E | A | K | V | K | E | Y | M | E | V | A | R | E | K | Y | K | E | R | D | D | R | M | R | G | L | T | D | E | T | V | D | T | F | Y | S | C | V | L | C | Q | S | F | A | P | N | H | V | C | I | V | T | P | E | R | V | G | L | C | G | A | V | S | W | L | D | A | K | A | S | Y | E | I | N | H | A | G | P | N | Q | P | I | P | K | E | G | E | I | D | P | I | K | G | I | W | K | S | V | N | D | Y | L | Y | T | A | S | N | R | N | L | E | Q | V | C | L | Y | T | L | M | E | N | P | M | T | S | C | G | C | F | E | - | A | I | M | A | I | L | P | E | C | N | G | I | M | I | T | T | R | D | H | A | G | M | T | P | S | G | M | T | F | S | T | L | A | G | M | I | G | G | G | T | Q | T | P | G | F | M | G | I | G | R | T | - | Y | I | V | S | K | K | F | I | S | A | D | G | G | I | A | R | I | V | W | M | P | - | - | - | - | K | S | L | K | D | F | L | H | D | E | F | V | R | R | S | V | E | E | G | L | G | E | D | F | I | D | K | I | A | D | E | T | I | G | T | T | V | D | E | I | L | P | Y | L | E | E | K | G | H | P | A | L | T | M | D | P | I | M |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1gnla_ | A | Q01770 | GO:0003824 |
| d1gnla_ | A | Q01770 | GO:0004601 |
| d1gnla_ | A | Q01770 | GO:0005737 |
| d1gnla_ | A | Q01770 | GO:0016491 |
| d1gnla_ | A | Q01770 | GO:0016661 |
| d1gnla_ | A | Q01770 | GO:0042542 |
| d1gnla_ | A | Q01770 | GO:0046872 |
| d1gnla_ | A | Q01770 | GO:0050418 |
| d1gnla_ | A | Q01770 | GO:0051536 |
| d1gnla_ | A | Q01770 | GO:0051539 |
| d1gnla_ | A | Q01770 | GO:0098869 |
| d1gnla_ | A | Q01770 | GO:0003824 |
| d1gnla_ | A | Q01770 | GO:0004601 |
| d1gnla_ | A | Q01770 | GO:0005737 |
| d1gnla_ | A | Q01770 | GO:0016491 |
| d1gnla_ | A | Q01770 | GO:0016661 |
| d1gnla_ | A | Q01770 | GO:0042542 |
| d1gnla_ | A | Q01770 | GO:0046872 |
| d1gnla_ | A | Q01770 | GO:0050418 |
| d1gnla_ | A | Q01770 | GO:0051536 |
| d1gnla_ | A | Q01770 | GO:0051539 |
| d1gnla_ | A | Q01770 | GO:0098869 |
