solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1rl4a- ( 66 ) kIvkypdpiLrrrSeeVt-----nfddnLkrvVr
d1v3ya- ( 1 ) mvy-pirlygdpvLrrkArpVe-----dfs-gIkrlAe
d1ws0a- ( 1 ) mavl-eiikhpnevLetpCerVi-----nfdkkLvklLk
d1xeoa- ( 1 ) svl-qVlhIpderLrkvAkpVe-----eVnaeIqriVd
d2os0a1 ( 1 ) mitmkdIiregnptLravAeeVpv----pIteeDrqlGe
d3e3ua- ( 2 ) avv-pIrivgdpvLhtaTtpVtvaadgslpadlaqlia
d3g5ka1 ( 6 ) f-s-hvCqvgdpvLrgvAapVe--raqLggpeLqrlTq
d3ocaa- ( 2 ) svl-sivtVpd-rLslcSeeVe------Vdqsir-lVd
d4dr8a- ( 3 ) aavairVakklappl-dlhyLgdrvLrqpAkrVs-----riddeLRqtIr
d4je6a1 ( 2 ) dlp-eivasgdpvLhekAreVd--pgeIgs-rIqkiId
d5hgwa1 ( 1 ) mir-eilkmgdprLlevAkpva-----fdtpeLheiVa
d6jf8a- ( 2 ) svvl-pVAkrgediLkliAapVsan--elnsnwLyqlAd
333 aaaaaa
60 70 80 90 100
d1rl4a- ( 95 ) kMfdiMyes------------kgiGLSAPQVniskRIIVwn---------
d1v3ya- ( 32 ) dMleTMfea------------kgvGLAAPQIGlsqrLFVAVelr------
d1ws0a- ( 34 ) DMheTMlia------------dgvGLAAPQVgvslqVAVVdvdd------
d1xeoa- ( 33 ) dMFeTMyae------------egiGLAATQVdihqrIIVIDvse------
d2os0a1 ( 36 ) dMltFLknSqdpvkaeelqLrgdvGLAAPQLdiskrIIAVhVp-------
d3e3ua- ( 39 ) tMydTMdaA------------ngvGLAANQIGcslrLFVYdCa-----ad
d3g5ka1 ( 40 ) rLvqvMrrr------------rcvGLSAPQLgvprqVLALeLp-ealCre
d3ocaa- ( 34 ) dMf-TMhan-------------glGLAAVQVgvhkrILVMnvpe------
d4dr8a- ( 49 ) qMLqTMySa------------dgiGLAAPQVginkqLIVIdlle------
d4je6a1 ( 37 ) dMikVMrla------------pcvGLAAPQIgvplrIIVLeDt-keyisy
d5hgwa1 ( 34 ) dMfeTMhha------------nGaGLAAPQIgiglqIIIFGfgsnnryp-
d6jf8a- ( 38 ) aMhaTMle------------rnGvGIAAPQVyiskrVIIVASr-------
aaaaaaaa bbb3333 bbbb
110 120 130 140 150
d1rl4a- ( 135 ) ----------------riFInPsiveqsl--vklkl---iegClSFp-gI
d1v3ya- ( 76 ) ------elvr----rvyvVAnPvityre---glve---gteg-lSLp-gl
d1ws0a- ( 66 ) ------d--t----gkieLINPsilekr---geqv---gpegclSfp-gl
d1xeoa- ( 65 ) --nrd---------erlVLInPelleks---getGi---eegClSIp-eq
d2os0a1 ( 88 ) -------------slstVMYNPkilshsv--qdVClge-gegCLSVdrdv
d3e3ua- ( 72 ) ram------tAr--rrgVVInPvletseipetmpdpdtddegClSVp-ge
d3g5ka1 ( 77 ) cpprqralrqmepfplrvFVnPslrvld--srlvt---fpegCeSva-gf
d3ocaa- ( 74 ) --eEdkiegyelygGpyCIInPkivdisq--ekvkl---kegClSVp-gy
d4dr8a- ( 82 ) ----------deqapplVLInPkiertag--dleqc---qegclSIp-gV
d4je6a1 ( 74 ) ap-eeilaqer-hfdlmVMVnPvlkers--nkkal---ffegCeSvd-gf
d5hgwa1 ( 71 ) ---------dAppVpeTvLInPkLeymp--pdmee---gwEGClSVp-gm
d6jf8a- ( 69 ) ---pnprypdApemnavvMVnPeIlefss--ecg-----eeGClSVp-de
bbbbbbbbbb bb bb
160 170 180 190 200
d1rl4a- ( 163 ) eg-kVeRpsiVsIsyydingykhlkilkgihSriFqhEfdHLnGtLFIdk
d1v3ya- ( 109 ) yseeVpRAerIrVeyqdeegrgrvlelegymArvFQhEidHLdGiLFFer
d1ws0a- ( 97 ) yG-eVeRadyIkVrAqnrrgkvflleAegflARaIQhEidHLhGvLFtsk
d1xeoa- ( 97 ) ra-lVpRaekVkIrAldrdgkpfeleAdgllAIcIqhEmdHLvGkLFMdy
d2os0a1 ( 122 ) pG-yVvRhNkItVSyfdmaGekhkvrlknyeAIVVqhEidHInGiMFydh
d3e3ua- ( 113 ) sf-pTgRAkwArVtgldadGspvsIegtglFArMLQhETGHLdGfLYldr
d3g5ka1 ( 121 ) lA-cVpRfqaVqIsgldpnGeqvvwqasgwaARIIQhEmdHLqGcLFidk
d3ocaa- ( 118 ) fd-yIvrpqrIaVqyldyngneciIkAQGwLARcLqhEidHLnGtvFLky
d4dr8a- ( 116 ) yL-dVeRpeiVeVsykdengp--qlvadgllArcIqhEmdHLnGvLFVDr
d4je6a1 ( 118 ) rA-AVeRyleVvVtgydrqgkrievnAsGwqARILqhEcdHLdGnLYvdk
d5hgwa1 ( 106 ) RG-VVsRyakVrYsgydqfgakidrvAegfhArVVQhEyDHLiGkLYpmr
d6jf8a- ( 110 ) rg-qVerae-VkVkyltlqG--vetvfqgfpAi-VQhEvdHLnGilFver
b b b bbbbbbb bbbbbb aaaaaaaaaaaaaa 333
210 220 230 240
d1rl4a- ( 212 ) Mt--------qvdkkkVrpkLnelird
d1v3ya- ( 159 ) Lp--kpk---reafleanraeLvrFqkea
d1ws0a- ( 146 ) Vtryye
d1xeoa- ( 146 ) ls---------------plkqqrirqkV--ekldrlk
d2os0a1 ( 171 ) In--------kenpfalk----------------egVlvie
d3e3ua- ( 162 ) L-i-gry---arnAkravkshgwgvpglswlPgedpdpfgh
d3g5ka1 ( 170 ) m-d-srtftnvywmkvnd
d3ocaa- ( 167 ) ls---------------kfkrdfAiekV--eli
d4dr8a- ( 165 ) Vn---------------rlelneaLdkk--gfavqavrpva
d4je6a1 ( 167 ) m-v-prTfrtvdnldlplae----------------gCplg
d5hgwa1 ( 155 ) Itd-ftrfgftevlfp-----------------------gl
d6jf8a- ( 159 ) is
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Peptide deformylase | Peptide deformylase | Malaria parasite (Plasmodium falciparum) [TaxId: 5833] | 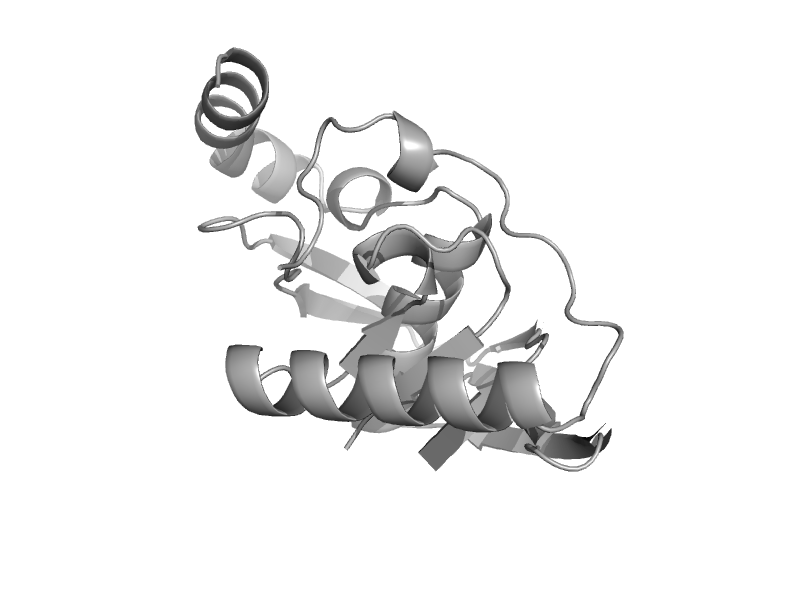 | view | |
| Peptide deformylase | Peptide deformylase | Thermus thermophilus [TaxId: 274] | 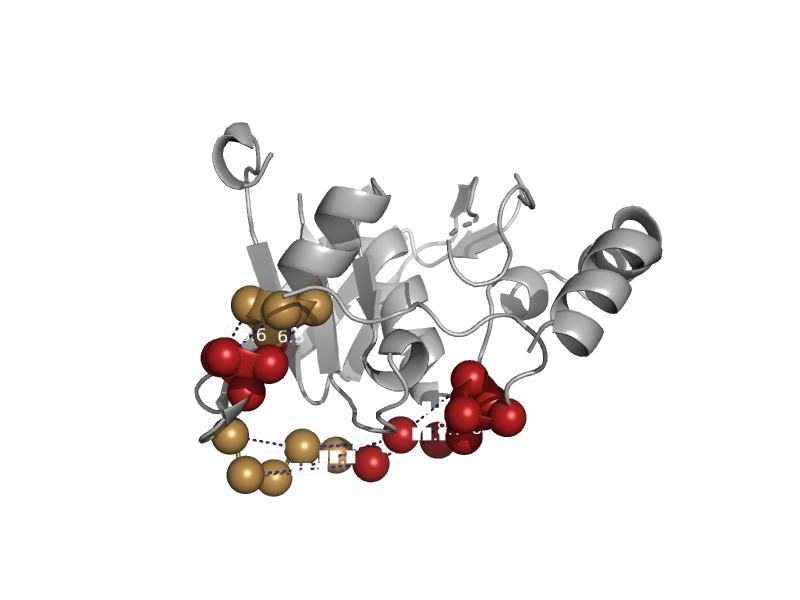 | view | |
| automated matches | automated matches | Bacillus cereus [TaxId: 1396] | 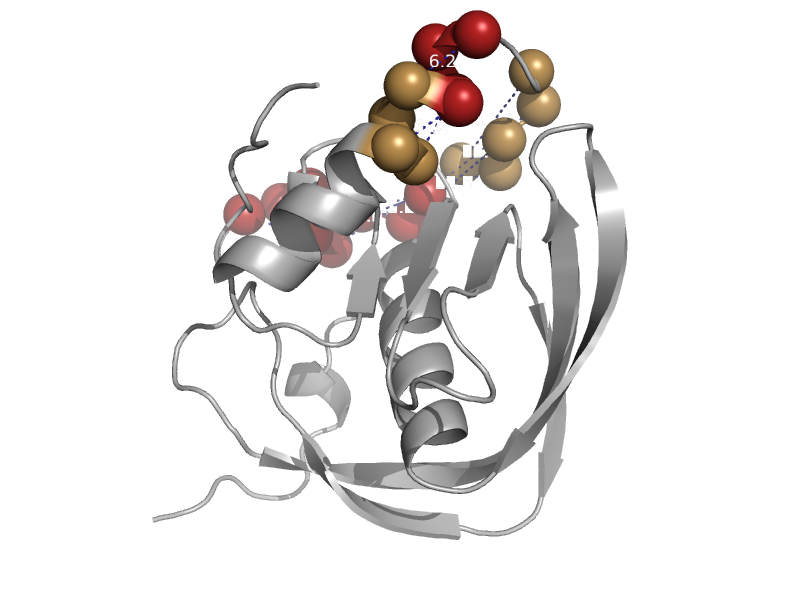 | view | |
| Peptide deformylase | Peptide deformylase | Escherichia coli [TaxId: 562] | 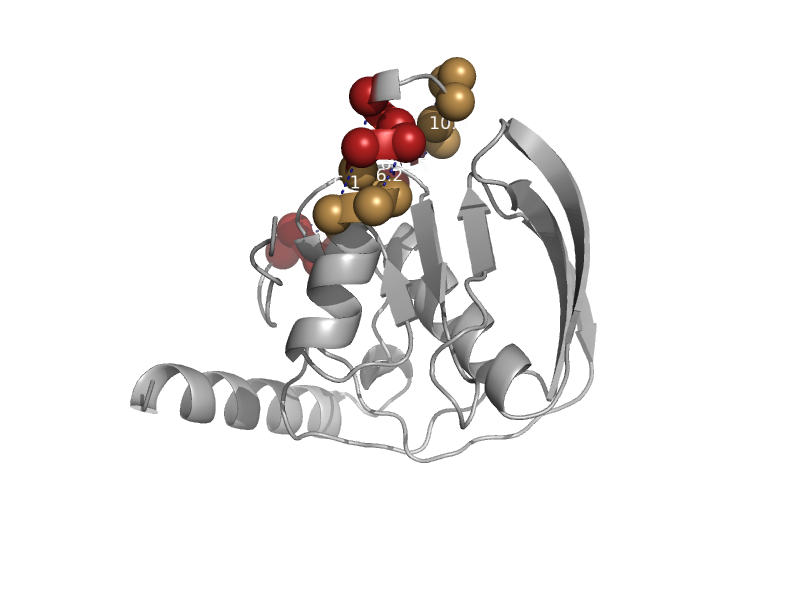 | view | |
| Peptide deformylase | Peptide deformylase | Enterococcus faecalis [TaxId: 1351] | 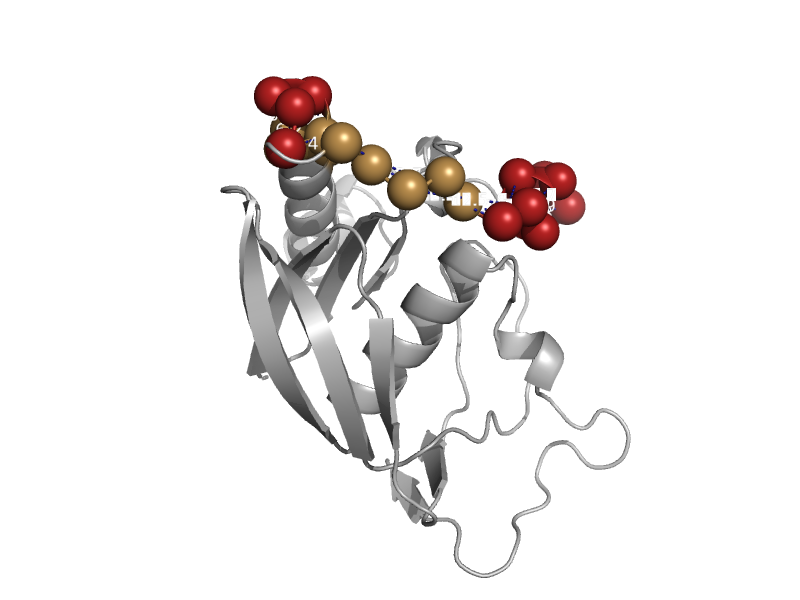 | view | |
| automated matches | automated matches | Mycobacterium tuberculosis [TaxId: 1773] | 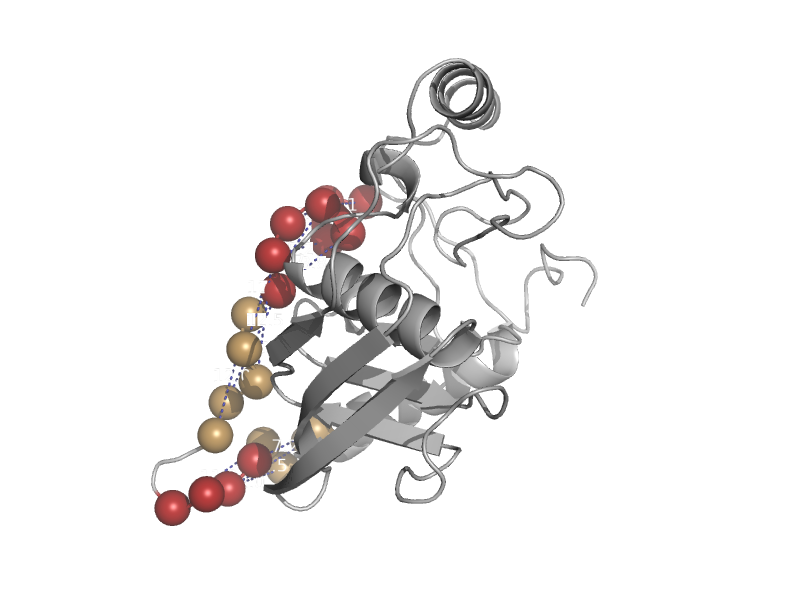 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 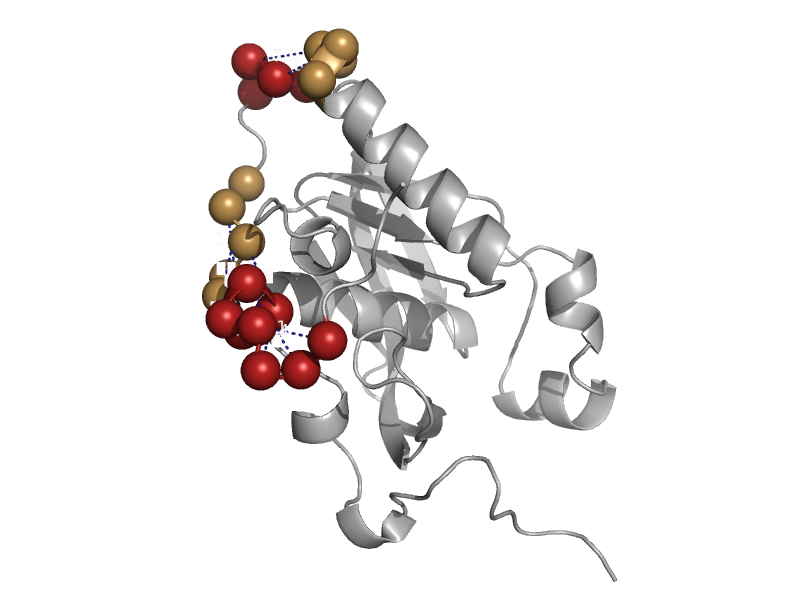 | view | |
| automated matches | automated matches | Ehrlichia chaffeensis [TaxId: 205920] | 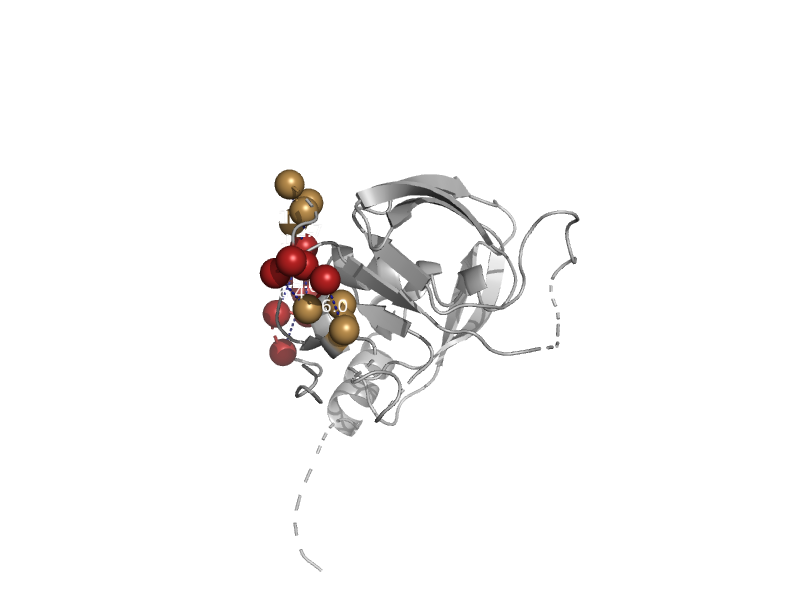 | view | |
| automated matches | automated matches | Synechococcus elongatus [TaxId: 269084] |  | view | |
| automated matches | automated matches | Thale cress (Arabidopsis thaliana) [TaxId: 3702] |  | view | |
| automated matches | automated matches | Burkholderia ambifaria [TaxId: 398577] |  | view | |
| automated matches | automated matches | Acinetobacter baumannii [TaxId: 470] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | I | V | K | Y | P | D | P | I | L | R | R | R | S | E | E | V | T | - | - | - | - | - | N | F | D | D | N | L | K | R | V | V | R | K | M | F | D | I | M | Y | E | S | - | - | - | - | - | - | - | - | - | - | - | - | K | G | I | G | L | S | A | P | Q | V | N | I | S | K | R | I | I | V | W | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | I | F | I | N | P | S | I | V | E | Q | S | L | - | - | V | K | L | K | L | - | - | - | I | E | G | C | L | S | F | P | - | G | I | E | G | - | K | V | E | R | P | S | I | V | S | I | S | Y | Y | D | I | N | G | Y | K | H | L | K | I | L | K | G | I | H | S | R | I | F | Q | H | E | F | D | H | L | N | G | T | L | F | I | D | K | M | T | - | - | - | - | - | - | - | - | Q | V | D | K | K | K | V | R | P | K | L | N | E | L | I | R | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | M | V | Y | - | P | I | R | L | Y | G | D | P | V | L | R | R | K | A | R | P | V | E | - | - | - | - | - | D | F | S | - | G | I | K | R | L | A | E | D | M | L | E | T | M | F | E | A | - | - | - | - | - | - | - | - | - | - | - | - | K | G | V | G | L | A | A | P | Q | I | G | L | S | Q | R | L | F | V | A | V | E | L | R | - | - | - | - | - | - | - | - | - | - | - | - | E | L | V | R | - | - | - | - | R | V | Y | V | V | A | N | P | V | I | T | Y | R | E | - | - | - | G | L | V | E | - | - | - | G | T | E | G | - | L | S | L | P | - | G | L | Y | S | E | E | V | P | R | A | E | R | I | R | V | E | Y | Q | D | E | E | G | R | G | R | V | L | E | L | E | G | Y | M | A | R | V | F | Q | H | E | I | D | H | L | D | G | I | L | F | F | E | R | L | P | - | - | K | P | K | - | - | - | R | E | A | F | L | E | A | N | R | A | E | L | V | R | F | Q | K | E | A | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | M | A | V | L | - | E | I | I | K | H | P | N | E | V | L | E | T | P | C | E | R | V | I | - | - | - | - | - | N | F | D | K | K | L | V | K | L | L | K | D | M | H | E | T | M | L | I | A | - | - | - | - | - | - | - | - | - | - | - | - | D | G | V | G | L | A | A | P | Q | V | G | V | S | L | Q | V | A | V | V | D | V | D | D | - | - | - | - | - | - | - | - | - | - | - | - | D | - | - | T | - | - | - | - | G | K | I | E | L | I | N | P | S | I | L | E | K | R | - | - | - | G | E | Q | V | - | - | - | G | P | E | G | C | L | S | F | P | - | G | L | Y | G | - | E | V | E | R | A | D | Y | I | K | V | R | A | Q | N | R | R | G | K | V | F | L | L | E | A | E | G | F | L | A | R | A | I | Q | H | E | I | D | H | L | H | G | V | L | F | T | S | K | V | T | R | Y | Y | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | S | V | L | - | Q | V | L | H | I | P | D | E | R | L | R | K | V | A | K | P | V | E | - | - | - | - | - | E | V | N | A | E | I | Q | R | I | V | D | D | M | F | E | T | M | Y | A | E | - | - | - | - | - | - | - | - | - | - | - | - | E | G | I | G | L | A | A | T | Q | V | D | I | H | Q | R | I | I | V | I | D | V | S | E | - | - | - | - | - | - | - | - | N | R | D | - | - | - | - | - | - | - | - | - | E | R | L | V | L | I | N | P | E | L | L | E | K | S | - | - | - | G | E | T | G | I | - | - | - | E | E | G | C | L | S | I | P | - | E | Q | R | A | - | L | V | P | R | A | E | K | V | K | I | R | A | L | D | R | D | G | K | P | F | E | L | E | A | D | G | L | L | A | I | C | I | Q | H | E | M | D | H | L | V | G | K | L | F | M | D | Y | L | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | L | K | Q | Q | R | I | R | Q | K | V | - | - | E | K | L | D | R | L | K | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | M | I | T | M | K | D | I | I | R | E | G | N | P | T | L | R | A | V | A | E | E | V | P | V | - | - | - | - | P | I | T | E | E | D | R | Q | L | G | E | D | M | L | T | F | L | K | N | S | Q | D | P | V | K | A | E | E | L | Q | L | R | G | D | V | G | L | A | A | P | Q | L | D | I | S | K | R | I | I | A | V | H | V | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | L | S | T | V | M | Y | N | P | K | I | L | S | H | S | V | - | - | Q | D | V | C | L | G | E | - | G | E | G | C | L | S | V | D | R | D | V | P | G | - | Y | V | V | R | H | N | K | I | T | V | S | Y | F | D | M | A | G | E | K | H | K | V | R | L | K | N | Y | E | A | I | V | V | Q | H | E | I | D | H | I | N | G | I | M | F | Y | D | H | I | N | - | - | - | - | - | - | - | - | K | E | N | P | F | A | L | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | G | V | L | V | I | E |
| - | - | - | - | - | - | - | - | - | - | - | - | A | V | V | - | P | I | R | I | V | G | D | P | V | L | H | T | A | T | T | P | V | T | V | A | A | D | G | S | L | P | A | D | L | A | Q | L | I | A | T | M | Y | D | T | M | D | A | A | - | - | - | - | - | - | - | - | - | - | - | - | N | G | V | G | L | A | A | N | Q | I | G | C | S | L | R | L | F | V | Y | D | C | A | - | - | - | - | - | A | D | R | A | M | - | - | - | - | - | - | T | A | R | - | - | R | R | G | V | V | I | N | P | V | L | E | T | S | E | I | P | E | T | M | P | D | P | D | T | D | D | E | G | C | L | S | V | P | - | G | E | S | F | - | P | T | G | R | A | K | W | A | R | V | T | G | L | D | A | D | G | S | P | V | S | I | E | G | T | G | L | F | A | R | M | L | Q | H | E | T | G | H | L | D | G | F | L | Y | L | D | R | L | - | I | - | G | R | Y | - | - | - | A | R | N | A | K | R | A | V | K | S | H | G | W | G | V | P | G | L | S | W | L | P | G | E | D | P | D | P | F | G | H |
| - | - | - | - | - | - | - | - | - | - | - | - | F | - | S | - | H | V | C | Q | V | G | D | P | V | L | R | G | V | A | A | P | V | E | - | - | R | A | Q | L | G | G | P | E | L | Q | R | L | T | Q | R | L | V | Q | V | M | R | R | R | - | - | - | - | - | - | - | - | - | - | - | - | R | C | V | G | L | S | A | P | Q | L | G | V | P | R | Q | V | L | A | L | E | L | P | - | E | A | L | C | R | E | C | P | P | R | Q | R | A | L | R | Q | M | E | P | F | P | L | R | V | F | V | N | P | S | L | R | V | L | D | - | - | S | R | L | V | T | - | - | - | F | P | E | G | C | E | S | V | A | - | G | F | L | A | - | C | V | P | R | F | Q | A | V | Q | I | S | G | L | D | P | N | G | E | Q | V | V | W | Q | A | S | G | W | A | A | R | I | I | Q | H | E | M | D | H | L | Q | G | C | L | F | I | D | K | M | - | D | - | S | R | T | F | T | N | V | Y | W | M | K | V | N | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | S | V | L | - | S | I | V | T | V | P | D | - | R | L | S | L | C | S | E | E | V | E | - | - | - | - | - | - | V | D | Q | S | I | R | - | L | V | D | D | M | F | - | T | M | H | A | N | - | - | - | - | - | - | - | - | - | - | - | - | - | G | L | G | L | A | A | V | Q | V | G | V | H | K | R | I | L | V | M | N | V | P | E | - | - | - | - | - | - | - | - | E | E | D | K | I | E | G | Y | E | L | Y | G | G | P | Y | C | I | I | N | P | K | I | V | D | I | S | Q | - | - | E | K | V | K | L | - | - | - | K | E | G | C | L | S | V | P | - | G | Y | F | D | - | Y | I | V | R | P | Q | R | I | A | V | Q | Y | L | D | Y | N | G | N | E | C | I | I | K | A | Q | G | W | L | A | R | C | L | Q | H | E | I | D | H | L | N | G | T | V | F | L | K | Y | L | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | F | K | R | D | F | A | I | E | K | V | - | - | E | L | I | - | - | - | - | - | - | - | - |
| A | A | V | A | I | R | V | A | K | K | L | A | P | P | L | - | D | L | H | Y | L | G | D | R | V | L | R | Q | P | A | K | R | V | S | - | - | - | - | - | R | I | D | D | E | L | R | Q | T | I | R | Q | M | L | Q | T | M | Y | S | A | - | - | - | - | - | - | - | - | - | - | - | - | D | G | I | G | L | A | A | P | Q | V | G | I | N | K | Q | L | I | V | I | D | L | L | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | E | Q | A | P | P | L | V | L | I | N | P | K | I | E | R | T | A | G | - | - | D | L | E | Q | C | - | - | - | Q | E | G | C | L | S | I | P | - | G | V | Y | L | - | D | V | E | R | P | E | I | V | E | V | S | Y | K | D | E | N | G | P | - | - | Q | L | V | A | D | G | L | L | A | R | C | I | Q | H | E | M | D | H | L | N | G | V | L | F | V | D | R | V | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | L | E | L | N | E | A | L | D | K | K | - | - | G | F | A | V | Q | A | V | R | P | V | A |
| - | - | - | - | - | - | - | - | - | - | - | - | D | L | P | - | E | I | V | A | S | G | D | P | V | L | H | E | K | A | R | E | V | D | - | - | P | G | E | I | G | S | - | R | I | Q | K | I | I | D | D | M | I | K | V | M | R | L | A | - | - | - | - | - | - | - | - | - | - | - | - | P | C | V | G | L | A | A | P | Q | I | G | V | P | L | R | I | I | V | L | E | D | T | - | K | E | Y | I | S | Y | A | P | - | E | E | I | L | A | Q | E | R | - | H | F | D | L | M | V | M | V | N | P | V | L | K | E | R | S | - | - | N | K | K | A | L | - | - | - | F | F | E | G | C | E | S | V | D | - | G | F | R | A | - | A | V | E | R | Y | L | E | V | V | V | T | G | Y | D | R | Q | G | K | R | I | E | V | N | A | S | G | W | Q | A | R | I | L | Q | H | E | C | D | H | L | D | G | N | L | Y | V | D | K | M | - | V | - | P | R | T | F | R | T | V | D | N | L | D | L | P | L | A | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | P | L | G |
| - | - | - | - | - | - | - | - | - | - | - | - | M | I | R | - | E | I | L | K | M | G | D | P | R | L | L | E | V | A | K | P | V | A | - | - | - | - | - | F | D | T | P | E | L | H | E | I | V | A | D | M | F | E | T | M | H | H | A | - | - | - | - | - | - | - | - | - | - | - | - | N | G | A | G | L | A | A | P | Q | I | G | I | G | L | Q | I | I | I | F | G | F | G | S | N | N | R | Y | P | - | - | - | - | - | - | - | - | - | - | D | A | P | P | V | P | E | T | V | L | I | N | P | K | L | E | Y | M | P | - | - | P | D | M | E | E | - | - | - | G | W | E | G | C | L | S | V | P | - | G | M | R | G | - | V | V | S | R | Y | A | K | V | R | Y | S | G | Y | D | Q | F | G | A | K | I | D | R | V | A | E | G | F | H | A | R | V | V | Q | H | E | Y | D | H | L | I | G | K | L | Y | P | M | R | I | T | D | - | F | T | R | F | G | F | T | E | V | L | F | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | L |
| - | - | - | - | - | - | - | - | - | - | - | S | V | V | L | - | P | V | A | K | R | G | E | D | I | L | K | L | I | A | A | P | V | S | A | N | - | - | E | L | N | S | N | W | L | Y | Q | L | A | D | A | M | H | A | T | M | L | E | - | - | - | - | - | - | - | - | - | - | - | - | R | N | G | V | G | I | A | A | P | Q | V | Y | I | S | K | R | V | I | I | V | A | S | R | - | - | - | - | - | - | - | - | - | - | P | N | P | R | Y | P | D | A | P | E | M | N | A | V | V | M | V | N | P | E | I | L | E | F | S | S | - | - | E | C | G | - | - | - | - | - | E | E | G | C | L | S | V | P | - | D | E | R | G | - | Q | V | E | R | A | E | - | V | K | V | K | Y | L | T | L | Q | G | - | - | V | E | T | V | F | Q | G | F | P | A | I | - | V | Q | H | E | V | D | H | L | N | G | I | L | F | V | E | R | I | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1rl4a_ | A | Q8I372 | GO:0042586 |
| d1rl4a_ | A | Q8I372 | GO:0042586 |
| d1v3ya_ | A | P43522 | GO:0006412 |
| d1v3ya_ | A | P43522 | GO:0016787 |
| d1v3ya_ | A | P43522 | GO:0018206 |
| d1v3ya_ | A | P43522 | GO:0042586 |
| d1v3ya_ | A | P43522 | GO:0046872 |
| d1v3ya_ | A | P43522 | GO:0006412 |
| d1v3ya_ | A | P43522 | GO:0016787 |
| d1v3ya_ | A | P43522 | GO:0018206 |
| d1v3ya_ | A | P43522 | GO:0042586 |
| d1v3ya_ | A | P43522 | GO:0046872 |
