solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1emsa2 ( 10 ) matgrhfIAVCQMtSd-n--dlekNFqaAknMIerAGekkCeMVFLPEcF
d1uf5a- ( 1 ) trqmiLAVGQQgpIaraetreqVVvrLldMLtkAasrgAnFIVFPELA
d2e11a- ( 1 ) mhdLrISLVQGsTrwh--dpagNrdyYgalLeplag-qSDLVILPETF
d2w1va- ( 41 ) tFrLALIQLqvs-s--iksdNLtrAcslVreAakqgAnIVSLPECF
d3ivza- ( 2 ) vkVAYVQMnPqil--epdkNyskAekLIkeAskqgAqLVVLPELF
d3p8ka1 ( 2 ) kVQIYQLpIvfG--dsskNetqItqwFeknna-eVdVVVLPE-W
d5h8ia- ( 4 ) dkgrkVvVSALQFaCtd---dvstNVttAerLVraAhkqgAnIVLIQELF
bbbbbbb aaaaaaaaaaaaaaaaa bbb
60 70 80 90 100
d1emsa2 ( 57 ) DFIGlnk------neqidlA--maTdceymekYreLArkhnIwLSLGGlh
d1uf5a- ( 49 ) LtTFFPRwhFtdeaeldsfYet-eMpgpvVrpLfekAaelgIGFN-LGYA
d2e11a- ( 46 ) TsGf-Sn------eaid-kaed--mdgpTVawIrtQAarLgAAIT-GSVQ
d2w1va- ( 84 ) NSpy-gt------tyFpdyA--ekIpgeSTqkLseVAkessIyLIGGSIP
d3ivza- ( 45 ) DTGynFe-tr---eeVfeiAqk-IpegeTTtfLmdvArdtgVyIV-AGTA
d3p8ka1 ( 44 ) nNgy-Dl------ehLnekADn--nLgqSfsfIkhLAekykVDIVAGSVs
d5h8ia- ( 51 ) egyYFCqAqr---edfiqrAkp-ykdHptImrLqkLAkelgVVIP-VSFF
aa aaaaaaaaaaaaa bbb bbbb
110 120 130 140 150
d1emsa2 ( 99 ) hkdp-sdaahpwnTHlIIdsdGvtraeYnKlhLfdleip-gkvrlm--es
d1uf5a- ( 97 ) eLvveggvkrrFNTSILVdksGkivgkYrKihLpghkeyeayrpfqhLEk
d2e11a- ( 85 ) Lrte----hgVfNRLLWATpdgal-qyYdKrhLf----r-fg---n--Eh
d2w1va- ( 125 ) Eeda-g---klyNTCSVFGpdGsllvkHrKihLfdidvp-gkitfq--Es
d3ivza- ( 89 ) Ekdg----dvLyNSAVVVGprg-figkYrKihLf------------yrEk
d3p8ka1 ( 85 ) Nirn----nqifnTAFSVnksGqlineYdKvhlv----p--l---r--Eh
d5h8ia- ( 96 ) Eean----nahyNSIAIIDadgtdlgiYrKshIpdg------pgye--Ek
bbb bbbbbbbbb bbbbbb 33
160 170 180 190 200
d1emsa2 ( 145 ) efskaGtemipPvdTp-IGrLGLSIcyDVrfpeLSlwNrk-r-----gAq
d1uf5a- ( 147 ) rYFepGdlgfpVydVd-aAkMGMFIanDRrwpeAWrvMGl-r-----gAe
d2e11a- ( 120 ) lrYaagr-erlCVewk-gWrINPQVcyDLrfpvfCrNrfdverpgqLDFD
d2w1va- ( 168 ) ktLspGdsfS-tFdTp-yckVGLGICyDMrfaeLAqIYAq-r-----gCq
d3ivza- ( 122 ) fFFepGdlgfrVfdLg-fMkVGVMI-fDWffpeSArtLAl-k-----gAD
d3p8ka1 ( 121 ) efLtaGeyvAepfqLsdgtyVTQLIcyDLrfpelLryPAr-s-----gAk
d5h8ia- ( 134 ) fYFnpGdtgfkvFqTk-yAkIGVAIcwDQwfpeAAraMAl-q-----gAe
3 bb bbbb 33333 aaaaaaaaa
210 220 230 240 250
d1emsa2 ( 188 ) LLSFPSAft---------lntGlahWetlLraRAieNQCYVVAAAQTgaH
d1uf5a- ( 190 ) IICGGYNtpthnPpvpqhdhltsfhHllsMqagSyqNGAWSAAAGKAgmE
d2e11a- ( 168 ) LQLFVANWp---------sara-yaWktlLraRAieNLCFVAAVNRVgvD
d2w1va- ( 210 ) LLVYPGaFn---------lttGpahWellQraRAvdNQVYVATASpArdd
d3ivza- ( 165 ) VIAHPANlvm-----p--------yApraMpirAleNkVYTVTADRVgeE
d3p8ka1 ( 165 ) IAFYVAQWp----------srl-qhWhslLkaRAienN-FVIGTNSTgfd
d5h8ia- ( 177 ) ILFYPTAiGs-----epqsidsrdhWkrvMqgHAgaNLVPLVASNRignE
bbbbb a aaaaaaaaaaaaaa bbbbb bb
260 270 280 290 300
d1emsa2 ( 229 ) np-------krqSyGhSMVVdPwGavvaqCse-rvdMcfaeIdLsyVdtl
d1uf5a- ( 240 ) e--------nCmLlGhSCIVAPtGeivalTttledeVitaaVdLdrcrel
d2e11a- ( 208 ) gn-------qlhYaGDSAVIdflGqpqveire-qeqvvtttIsaaaLaeh
d2w1va- ( 251 ) ka-------sYvawGhSTVVDPwgqvltkagt-eetilysdIdlkkLaeI
d3ivza- ( 202 ) r--------glkFiGkSLIASPkaevlsmAseteeevgVaeIdlslVrn-
d3p8ka1 ( 205 ) g--------nteYaGhSiVInPnGdlvgelne-sadiltVdLnlneVeqq
d5h8ia- ( 224 ) iieTehgkseikFYGnSFIAgptgeivsiaddkeeavliaefnLdkiksm
bb bbb bbbb bbbbbbb aaaaaaa
310 320 330
d1emsa2 ( 271 ) re----mqpvfshr
d1uf5a- ( 282 ) reh---ifnFkqhrqpqhygliael
d2e11a- ( 250 ) ra----rfpamldgd------------sfvlg
d2w1va- ( 293 ) rq----qipilkqkrad-------lytves
d3ivza- ( 243 ) -krindlndifkdrreeyyfr
d3p8ka1 ( 246 ) re----nipvfksikld-------lyk
d5h8ia- ( 274 ) rh---cw-GvfrdrrpdlykvllTldgknpvl
aa a
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| NIT-FHIT fusion protein, N-terminal domain | Nitrilase | Nematode (Caenorhabditis elegans) [TaxId: 6239] | 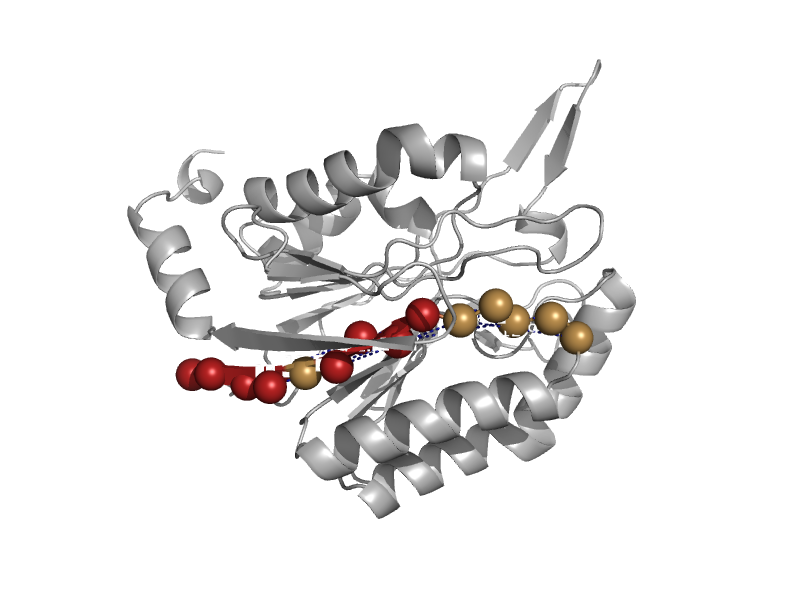 | view | |
| N-carbamoyl-D-aminoacid amidohydrolase | Carbamilase | Agrobacterium sp. [TaxId: 361] | 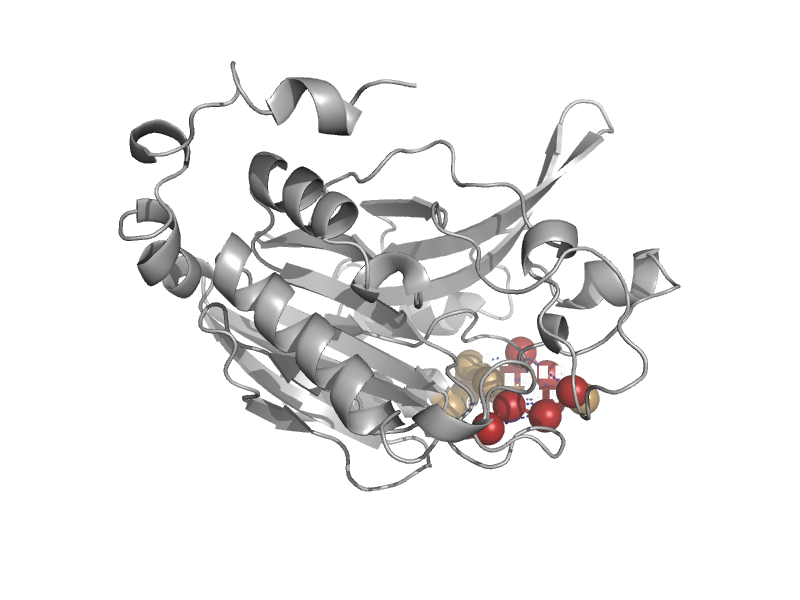 | view | |
| automated matches | automated matches | Xanthomonas campestris pv. campestris [TaxId: 340] | 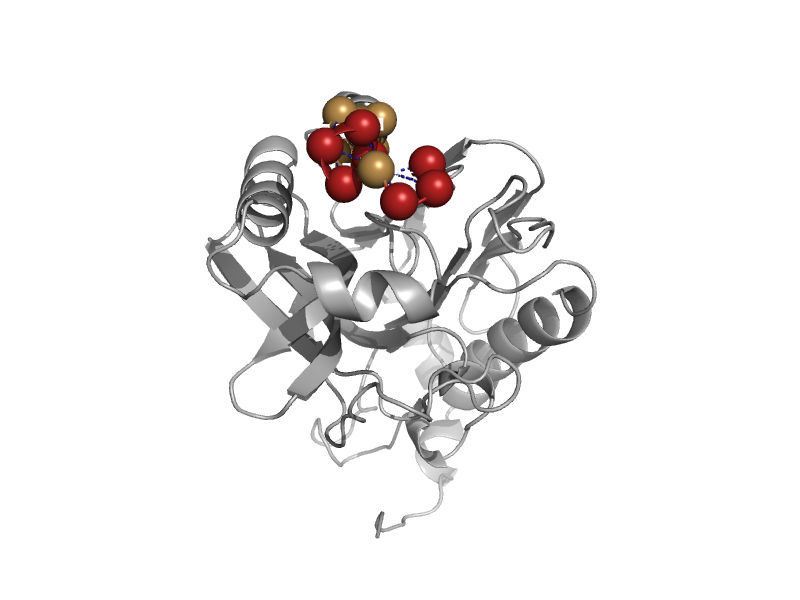 | view | |
| automated matches | automated matches | Mouse (Mus musculus) [TaxId: 10090] | 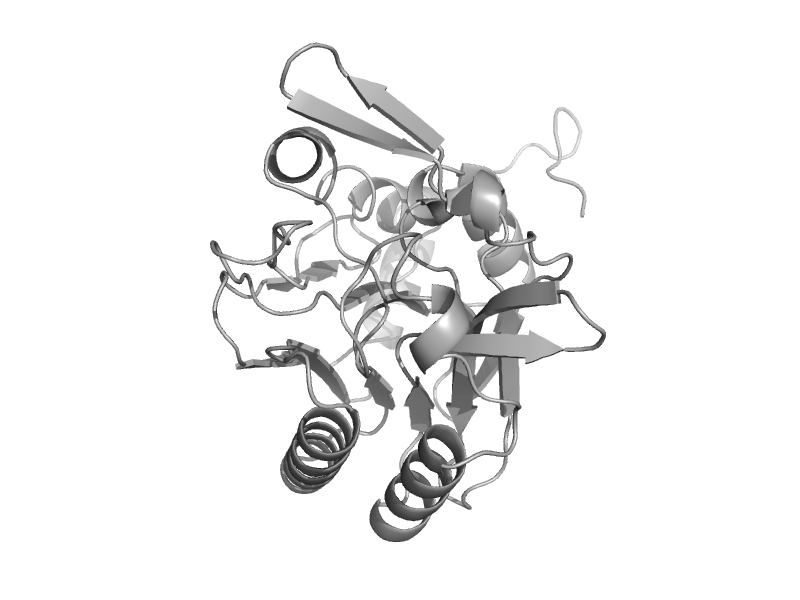 | view | |
| automated matches | Carbamilase | Pyrococcus abyssi [TaxId: 272844] |  | view | |
| automated matches | automated matches | Staphylococcus aureus [TaxId: 93062] |  | view | |
| automated matches | automated matches | Medicago truncatula [TaxId: 3880] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | 330 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | A | T | G | R | H | F | I | A | V | C | Q | M | T | S | D | - | N | - | - | D | L | E | K | N | F | Q | A | A | K | N | M | I | E | R | A | G | E | K | K | C | E | M | V | F | L | P | E | C | F | D | F | I | G | L | N | K | - | - | - | - | - | - | N | E | Q | I | D | L | A | - | - | M | A | T | D | C | E | Y | M | E | K | Y | R | E | L | A | R | K | H | N | I | W | L | S | L | G | G | L | H | H | K | D | P | - | S | D | A | A | H | P | W | N | T | H | L | I | I | D | S | D | G | V | T | R | A | E | Y | N | K | L | H | L | F | D | L | E | I | P | - | G | K | V | R | L | M | - | - | E | S | E | F | S | K | A | G | T | E | M | I | P | P | V | D | T | P | - | I | G | R | L | G | L | S | I | C | Y | D | V | R | F | P | E | L | S | L | W | N | R | K | - | R | - | - | - | - | - | G | A | Q | L | L | S | F | P | S | A | F | T | - | - | - | - | - | - | - | - | - | L | N | T | G | L | A | H | W | E | T | L | L | R | A | R | A | I | E | N | Q | C | Y | V | V | A | A | A | Q | T | G | A | H | N | P | - | - | - | - | - | - | - | K | R | Q | S | Y | G | H | S | M | V | V | D | P | W | G | A | V | V | A | Q | C | S | E | - | R | V | D | M | C | F | A | E | I | D | L | S | Y | V | D | T | L | R | E | - | - | - | - | M | Q | P | V | F | S | H | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | T | R | Q | M | I | L | A | V | G | Q | Q | G | P | I | A | R | A | E | T | R | E | Q | V | V | V | R | L | L | D | M | L | T | K | A | A | S | R | G | A | N | F | I | V | F | P | E | L | A | L | T | T | F | F | P | R | W | H | F | T | D | E | A | E | L | D | S | F | Y | E | T | - | E | M | P | G | P | V | V | R | P | L | F | E | K | A | A | E | L | G | I | G | F | N | - | L | G | Y | A | E | L | V | V | E | G | G | V | K | R | R | F | N | T | S | I | L | V | D | K | S | G | K | I | V | G | K | Y | R | K | I | H | L | P | G | H | K | E | Y | E | A | Y | R | P | F | Q | H | L | E | K | R | Y | F | E | P | G | D | L | G | F | P | V | Y | D | V | D | - | A | A | K | M | G | M | F | I | A | N | D | R | R | W | P | E | A | W | R | V | M | G | L | - | R | - | - | - | - | - | G | A | E | I | I | C | G | G | Y | N | T | P | T | H | N | P | P | V | P | Q | H | D | H | L | T | S | F | H | H | L | L | S | M | Q | A | G | S | Y | Q | N | G | A | W | S | A | A | A | G | K | A | G | M | E | E | - | - | - | - | - | - | - | - | N | C | M | L | L | G | H | S | C | I | V | A | P | T | G | E | I | V | A | L | T | T | T | L | E | D | E | V | I | T | A | A | V | D | L | D | R | C | R | E | L | R | E | H | - | - | - | I | F | N | F | K | Q | H | R | Q | P | Q | H | Y | G | L | I | A | E | L | - | - | - | - | - | - | - |
| - | - | M | H | D | L | R | I | S | L | V | Q | G | S | T | R | W | H | - | - | D | P | A | G | N | R | D | Y | Y | G | A | L | L | E | P | L | A | G | - | Q | S | D | L | V | I | L | P | E | T | F | T | S | G | F | - | S | N | - | - | - | - | - | - | E | A | I | D | - | K | A | E | D | - | - | M | D | G | P | T | V | A | W | I | R | T | Q | A | A | R | L | G | A | A | I | T | - | G | S | V | Q | L | R | T | E | - | - | - | - | H | G | V | F | N | R | L | L | W | A | T | P | D | G | A | L | - | Q | Y | Y | D | K | R | H | L | F | - | - | - | - | R | - | F | G | - | - | - | N | - | - | E | H | L | R | Y | A | A | G | R | - | E | R | L | C | V | E | W | K | - | G | W | R | I | N | P | Q | V | C | Y | D | L | R | F | P | V | F | C | R | N | R | F | D | V | E | R | P | G | Q | L | D | F | D | L | Q | L | F | V | A | N | W | P | - | - | - | - | - | - | - | - | - | S | A | R | A | - | Y | A | W | K | T | L | L | R | A | R | A | I | E | N | L | C | F | V | A | A | V | N | R | V | G | V | D | G | N | - | - | - | - | - | - | - | Q | L | H | Y | A | G | D | S | A | V | I | D | F | L | G | Q | P | Q | V | E | I | R | E | - | Q | E | Q | V | V | T | T | T | I | S | A | A | A | L | A | E | H | R | A | - | - | - | - | R | F | P | A | M | L | D | G | D | - | - | - | - | - | - | - | - | - | - | - | - | S | F | V | L | G |
| - | - | - | - | T | F | R | L | A | L | I | Q | L | Q | V | S | - | S | - | - | I | K | S | D | N | L | T | R | A | C | S | L | V | R | E | A | A | K | Q | G | A | N | I | V | S | L | P | E | C | F | N | S | P | Y | - | G | T | - | - | - | - | - | - | T | Y | F | P | D | Y | A | - | - | E | K | I | P | G | E | S | T | Q | K | L | S | E | V | A | K | E | S | S | I | Y | L | I | G | G | S | I | P | E | E | D | A | - | G | - | - | - | K | L | Y | N | T | C | S | V | F | G | P | D | G | S | L | L | V | K | H | R | K | I | H | L | F | D | I | D | V | P | - | G | K | I | T | F | Q | - | - | E | S | K | T | L | S | P | G | D | S | F | S | - | T | F | D | T | P | - | Y | C | K | V | G | L | G | I | C | Y | D | M | R | F | A | E | L | A | Q | I | Y | A | Q | - | R | - | - | - | - | - | G | C | Q | L | L | V | Y | P | G | A | F | N | - | - | - | - | - | - | - | - | - | L | T | T | G | P | A | H | W | E | L | L | Q | R | A | R | A | V | D | N | Q | V | Y | V | A | T | A | S | P | A | R | D | D | K | A | - | - | - | - | - | - | - | S | Y | V | A | W | G | H | S | T | V | V | D | P | W | G | Q | V | L | T | K | A | G | T | - | E | E | T | I | L | Y | S | D | I | D | L | K | K | L | A | E | I | R | Q | - | - | - | - | Q | I | P | I | L | K | Q | K | R | A | D | - | - | - | - | - | - | - | L | Y | T | V | E | S | - | - |
| - | - | - | - | - | V | K | V | A | Y | V | Q | M | N | P | Q | I | L | - | - | E | P | D | K | N | Y | S | K | A | E | K | L | I | K | E | A | S | K | Q | G | A | Q | L | V | V | L | P | E | L | F | D | T | G | Y | N | F | E | - | T | R | - | - | - | E | E | V | F | E | I | A | Q | K | - | I | P | E | G | E | T | T | T | F | L | M | D | V | A | R | D | T | G | V | Y | I | V | - | A | G | T | A | E | K | D | G | - | - | - | - | D | V | L | Y | N | S | A | V | V | V | G | P | R | G | - | F | I | G | K | Y | R | K | I | H | L | F | - | - | - | - | - | - | - | - | - | - | - | - | Y | R | E | K | F | F | F | E | P | G | D | L | G | F | R | V | F | D | L | G | - | F | M | K | V | G | V | M | I | - | F | D | W | F | F | P | E | S | A | R | T | L | A | L | - | K | - | - | - | - | - | G | A | D | V | I | A | H | P | A | N | L | V | M | - | - | - | - | - | P | - | - | - | - | - | - | - | - | Y | A | P | R | A | M | P | I | R | A | L | E | N | K | V | Y | T | V | T | A | D | R | V | G | E | E | R | - | - | - | - | - | - | - | - | G | L | K | F | I | G | K | S | L | I | A | S | P | K | A | E | V | L | S | M | A | S | E | T | E | E | E | V | G | V | A | E | I | D | L | S | L | V | R | N | - | - | K | R | I | N | D | L | N | D | I | F | K | D | R | R | E | E | Y | Y | F | R | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | K | V | Q | I | Y | Q | L | P | I | V | F | G | - | - | D | S | S | K | N | E | T | Q | I | T | Q | W | F | E | K | N | N | A | - | E | V | D | V | V | V | L | P | E | - | W | N | N | G | Y | - | D | L | - | - | - | - | - | - | E | H | L | N | E | K | A | D | N | - | - | N | L | G | Q | S | F | S | F | I | K | H | L | A | E | K | Y | K | V | D | I | V | A | G | S | V | S | N | I | R | N | - | - | - | - | N | Q | I | F | N | T | A | F | S | V | N | K | S | G | Q | L | I | N | E | Y | D | K | V | H | L | V | - | - | - | - | P | - | - | L | - | - | - | R | - | - | E | H | E | F | L | T | A | G | E | Y | V | A | E | P | F | Q | L | S | D | G | T | Y | V | T | Q | L | I | C | Y | D | L | R | F | P | E | L | L | R | Y | P | A | R | - | S | - | - | - | - | - | G | A | K | I | A | F | Y | V | A | Q | W | P | - | - | - | - | - | - | - | - | - | - | S | R | L | - | Q | H | W | H | S | L | L | K | A | R | A | I | E | N | N | - | F | V | I | G | T | N | S | T | G | F | D | G | - | - | - | - | - | - | - | - | N | T | E | Y | A | G | H | S | I | V | I | N | P | N | G | D | L | V | G | E | L | N | E | - | S | A | D | I | L | T | V | D | L | N | L | N | E | V | E | Q | Q | R | E | - | - | - | - | N | I | P | V | F | K | S | I | K | L | D | - | - | - | - | - | - | - | L | Y | K | - | - | - | - | - |
| D | K | G | R | K | V | V | V | S | A | L | Q | F | A | C | T | D | - | - | - | D | V | S | T | N | V | T | T | A | E | R | L | V | R | A | A | H | K | Q | G | A | N | I | V | L | I | Q | E | L | F | E | G | Y | Y | F | C | Q | A | Q | R | - | - | - | E | D | F | I | Q | R | A | K | P | - | Y | K | D | H | P | T | I | M | R | L | Q | K | L | A | K | E | L | G | V | V | I | P | - | V | S | F | F | E | E | A | N | - | - | - | - | N | A | H | Y | N | S | I | A | I | I | D | A | D | G | T | D | L | G | I | Y | R | K | S | H | I | P | D | G | - | - | - | - | - | - | P | G | Y | E | - | - | E | K | F | Y | F | N | P | G | D | T | G | F | K | V | F | Q | T | K | - | Y | A | K | I | G | V | A | I | C | W | D | Q | W | F | P | E | A | A | R | A | M | A | L | - | Q | - | - | - | - | - | G | A | E | I | L | F | Y | P | T | A | I | G | S | - | - | - | - | - | E | P | Q | S | I | D | S | R | D | H | W | K | R | V | M | Q | G | H | A | G | A | N | L | V | P | L | V | A | S | N | R | I | G | N | E | I | I | E | T | E | H | G | K | S | E | I | K | F | Y | G | N | S | F | I | A | G | P | T | G | E | I | V | S | I | A | D | D | K | E | E | A | V | L | I | A | E | F | N | L | D | K | I | K | S | M | R | H | - | - | - | C | W | - | G | V | F | R | D | R | R | P | D | L | Y | K | V | L | L | T | L | D | G | K | N | P | V | L |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1emsa2 | A | O76463 | GO:0000166 |
| d1emsa2 | A | O76463 | GO:0003824 |
| d1emsa2 | A | O76463 | GO:0005575 |
| d1emsa2 | A | O76463 | GO:0006139 |
| d1emsa2 | A | O76463 | GO:0016787 |
| d1emsa2 | A | O76463 | GO:0016811 |
| d1emsa2 | A | O76463 | GO:0043605 |
| d1emsa2 | A | O76463 | GO:0047710 |
| d1emsa2 | A | O76463 | GO:0110050 |
| d1emsa2 | A | O76463 | GO:0000166 |
| d1emsa2 | A | O76463 | GO:0003824 |
| d1emsa2 | A | O76463 | GO:0005575 |
| d1emsa2 | A | O76463 | GO:0006139 |
| d1emsa2 | A | O76463 | GO:0016787 |
| d1emsa2 | A | O76463 | GO:0016811 |
| d1emsa2 | A | O76463 | GO:0043605 |
| d1emsa2 | A | O76463 | GO:0047710 |
| d1emsa2 | A | O76463 | GO:0110050 |
| d1uf5a_ | A | P60327 | GO:0003837 |
| d1uf5a_ | A | P60327 | GO:0016787 |
| d1uf5a_ | A | P60327 | GO:0033396 |
| d1uf5a_ | A | P60327 | GO:0047417 |
| d1uf5a_ | A | P60327 | GO:0003837 |
| d1uf5a_ | A | P60327 | GO:0016787 |
| d1uf5a_ | A | P60327 | GO:0033396 |
| d1uf5a_ | A | P60327 | GO:0047417 |
