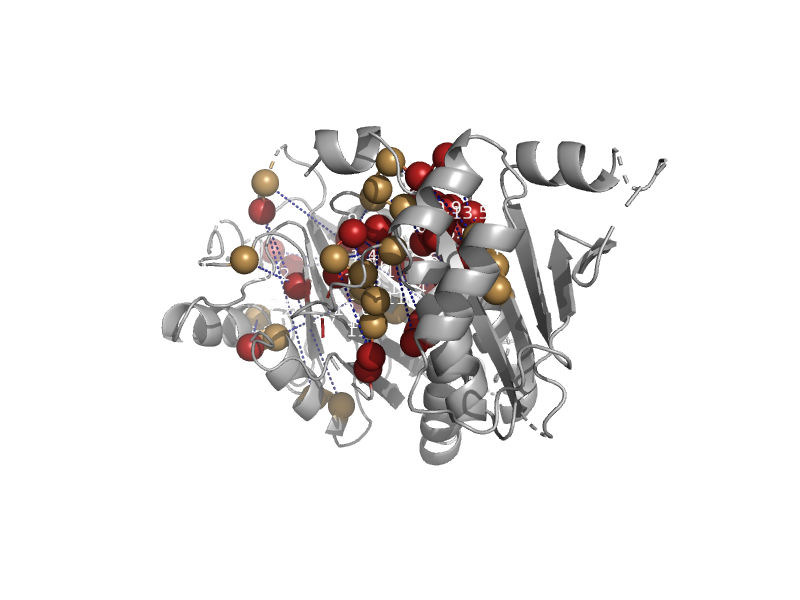
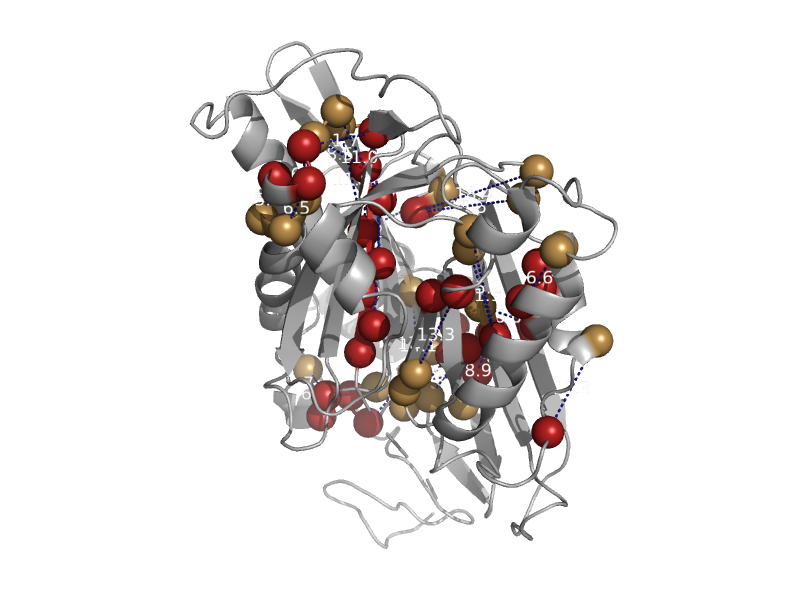
solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1vz6a- ( 8 ) tPrGFVVhTapVgLad---------grd
d3ndva- ( 1 ) gprArdlgVpFegtpgalnAITdVagVeVghttvisgdgamvigkGPyRT
bbbbb
60 70 80 90 100
d1vz6a- ( 28 ) DFTVLASt-----apAtVSAVFTrsrfagpsVvlCreAvadgqArGVVVL
d3ndva- ( 51 ) GVTIIHPlgktsldGVAAGravingtgeWtgMhlVde---vgqFlgPIAL
bbbbb bbbbbb aaaaaa bbb
110 120 130 140 150
d1vz6a- ( 73 ) ArnANVaTgleGeeNAreVraVaaL---gLpegeMLIASTgv---igqYp
d3ndva- ( 98 ) TG-tgnv--glVhqSMmdWSvgkVpeealfsrllPVVAeTlDnrLNDvfG
bb aaaaaaaaaa 3 bbbbb
160 170 180 190 200
d1vz6a- ( 120 ) msIrhLtLwpa----geggFrAAr-AImttDt--------pKevvvg---
d3ndva- ( 145 ) hgLtrdhVfaALdgAkggpVaEGnvGGGTGMiAYtFKGGIGTSSRvvsAg
bbb
210 220 230 240 250
d1vz6a- ( 162 ) --gAtLvGIAKGVGmL--ePdMa---------------------------
d3ndva- ( 195 ) dtrytVGVLVQAnHgdRndLrIagvqIGkeIkgawpevngivaagpdagk
bbbbbb
260 270 280 290 300
d1vz6a- ( 181 ) -TLLTFFATDArLdpaeQdrLFrrVMdrTFnAVSIdtdtStSDTAVLFAN
d3ndva- ( 245 ) psLLIVIATdAPLmphQLerMAr-rAalGVgrNgStagalsGEfALAFST
bbbbbb aaaaaaaaa aaa bbbbbbb
310 320 330 340 350
d1vz6a- ( 230 ) glagvd-----------------ageFe-aLhtAAlaLVkdIAsdGegAa
d3ndva- ( 298 ) shViplggkprlpaiindtdsetMnALfrGVvqATeEALVNQLVaSetmt
bbbb aaaa aaaaaaaaaaaaaa
360 370 380 390 400
d1vz6a- ( 264 ) kLIeVqVtgArddaQAkrVGktVVnSplVktaVhgcdpnwgrVamAIgkc
d3ndva- ( 348 ) GAnnakVygIp-hdqLarIMkarfp
bbb aaaaaaaaaaa
410 420 430 440 450
d1vz6a- ( 314 ) sdDtdidqerVtIrFgevevyppalaavaehLrgdeVvIGIdLaiagaft
d3ndva-
460
d1vz6a- ( 375 ) VYGCDltegyvrln
d3ndva-
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | 330 | 340 | 350 | 360 | 370 | 380 | 390 | 400 | 410 | 420 | 430 | 440 | 450 | 460 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | P | R | G | F | V | V | H | T | A | P | V | G | L | A | D | - | - | - | - | - | - | - | - | - | G | R | D | D | F | T | V | L | A | S | T | - | - | - | - | - | A | P | A | T | V | S | A | V | F | T | R | S | R | F | A | G | P | S | V | V | L | C | R | E | A | V | A | D | G | Q | A | R | G | V | V | V | L | A | R | N | A | N | V | A | T | G | L | E | G | E | E | N | A | R | E | V | R | A | V | A | A | L | - | - | - | G | L | P | E | G | E | M | L | I | A | S | T | G | V | - | - | - | I | G | Q | Y | P | M | S | I | R | H | L | T | L | W | P | A | - | - | - | - | G | E | G | G | F | R | A | A | R | - | A | I | M | T | T | D | T | - | - | - | - | - | - | - | - | P | K | E | V | V | V | G | - | - | - | - | - | G | A | T | L | V | G | I | A | K | G | V | G | M | L | - | - | E | P | D | M | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | L | L | T | F | F | A | T | D | A | R | L | D | P | A | E | Q | D | R | L | F | R | R | V | M | D | R | T | F | N | A | V | S | I | D | T | D | T | S | T | S | D | T | A | V | L | F | A | N | G | L | A | G | V | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | G | E | F | E | - | A | L | H | T | A | A | L | A | L | V | K | D | I | A | S | D | G | E | G | A | A | K | L | I | E | V | Q | V | T | G | A | R | D | D | A | Q | A | K | R | V | G | K | T | V | V | N | S | P | L | V | K | T | A | V | H | G | C | D | P | N | W | G | R | V | A | M | A | I | G | K | C | S | D | D | T | D | I | D | Q | E | R | V | T | I | R | F | G | E | V | E | V | Y | P | P | A | L | A | A | V | A | E | H | L | R | G | D | E | V | V | I | G | I | D | L | A | I | A | G | A | F | T | V | Y | G | C | D | L | T | E | G | Y | V | R | L | N |
| G | P | R | A | R | D | L | G | V | P | F | E | G | T | P | G | A | L | N | A | I | T | D | V | A | G | V | E | V | G | H | T | T | V | I | S | G | D | G | A | M | V | I | G | K | G | P | Y | R | T | G | V | T | I | I | H | P | L | G | K | T | S | L | D | G | V | A | A | G | R | A | V | I | N | G | T | G | E | W | T | G | M | H | L | V | D | E | - | - | - | V | G | Q | F | L | G | P | I | A | L | T | G | - | T | G | N | V | - | - | G | L | V | H | Q | S | M | M | D | W | S | V | G | K | V | P | E | E | A | L | F | S | R | L | L | P | V | V | A | E | T | L | D | N | R | L | N | D | V | F | G | H | G | L | T | R | D | H | V | F | A | A | L | D | G | A | K | G | G | P | V | A | E | G | N | V | G | G | G | T | G | M | I | A | Y | T | F | K | G | G | I | G | T | S | S | R | V | V | S | A | G | D | T | R | Y | T | V | G | V | L | V | Q | A | N | H | G | D | R | N | D | L | R | I | A | G | V | Q | I | G | K | E | I | K | G | A | W | P | E | V | N | G | I | V | A | A | G | P | D | A | G | K | P | S | L | L | I | V | I | A | T | D | A | P | L | M | P | H | Q | L | E | R | M | A | R | - | R | A | A | L | G | V | G | R | N | G | S | T | A | G | A | L | S | G | E | F | A | L | A | F | S | T | S | H | V | I | P | L | G | G | K | P | R | L | P | A | I | I | N | D | T | D | S | E | T | M | N | A | L | F | R | G | V | V | Q | A | T | E | E | A | L | V | N | Q | L | V | A | S | E | T | M | T | G | A | N | N | A | K | V | Y | G | I | P | - | H | D | Q | L | A | R | I | M | K | A | R | F | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |