solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d2wasa- (1767 ) g--gvgvdvelitsInven----dtfiernFtpqEieyCsaqp---svq
d2wdsa- ( 1 ) msiigvgidvaeverfgaalert-palagrLFlesElllpgger---rgv
d3hqja- ( 2 ) givgvgidlvsipdfaeqv----d-----tftpgErrdAs------saa
d3otaa1 ( 2 ) ivglgtdiaeIervekalarsgenfArrILtdsEleqFhask---qqG
d4mrta1 ( 1 ) mkiygiymdrpls----qeenerfmtfIspekrekCrrfyhkedaH
d4mrta2 ( 102 ) qpigidiektkpis-------leiAkrfFsktEysdLlakdd--eqt
d5xuka- ( 1 ) migidiVsiarIekcvkrfkmkflerFLspsEivlCkd------ks
bbbbbbb aaaaaa aaaaaaa aa
60 70 80 90 100
d2wasa- (1807 ) ssFAGTwSAKeAVfkSLg--------valkdIeIvrvnknapaVelhgna
d2wdsa- ( 47 ) aslAaRfAAKeALakALgap--a--gllwtdAeVwveaggrprLrvtgtV
d3hqja- ( 48 ) rhlAArwAAKeAVikAwsgsrfaqrpmihrdIeVvtdmwgrprVrltgaI
d3otaa1 ( 47 ) rfLAkRfAAKeAASkALgtgiaq--gVtfhdFtIshdklgkplLiLsgqA
d4mrta1 ( 43 ) rtLlGdvlVRsvISrqyq--------ldksdIfst-qeygkpcIpd----
d4mrta2 ( 141 ) dYFYhLwSMKeSFIkqegk----glslpldsFsVrlhqdgqVsIel----
d5xuka- ( 41 ) ssIAGFfALKeACSkALqvgigk--eLsfldIkIskspknaplItLs---
aaaaaaaaaaaaaaaa 333bbbbb bbbbb
110 120 130 140
d2wasa- (1856 ) kkaAeeagV-dVkVsIsh-ddlqAvAvAvStk
d2wdsa- ( 93 ) aarAaelgVaswhVslsa-dagiAsAvViAeg
d3hqja- ( 102 ) aeyLa--dv-tIhVslth-egdtAaAvAiLeap
d3otaa1 ( 95 ) aelasqlqVenihLsisd-erhyA-AtViLer
d4mrta1 ( 81 ) lpda------hFnishs---grwViGaFds
d4mrta2 ( 183 ) pshs------pCyIktyevdpgykmAvCAahpdFpeditmvsyeell
d5xuka- ( 86 ) kekmdyfnIqsLsAsish-dagfAiAvVvVs
aaa bbbbbb bbbbbbbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| automated matches | automated matches | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] |  | view | |
| automated matches | automated matches | Streptomyces coelicolor [TaxId: 1902] |  | view | |
| automated matches | automated matches | Mycobacterium tuberculosis [TaxId: 1773] |  | view | |
| automated matches | automated matches | Vibrio cholerae [TaxId: 243277] |  | view | |
| automated matches | 4'-Phosphopantetheinyl transferase SFP | Bacillus subtilis [TaxId: 224308] | 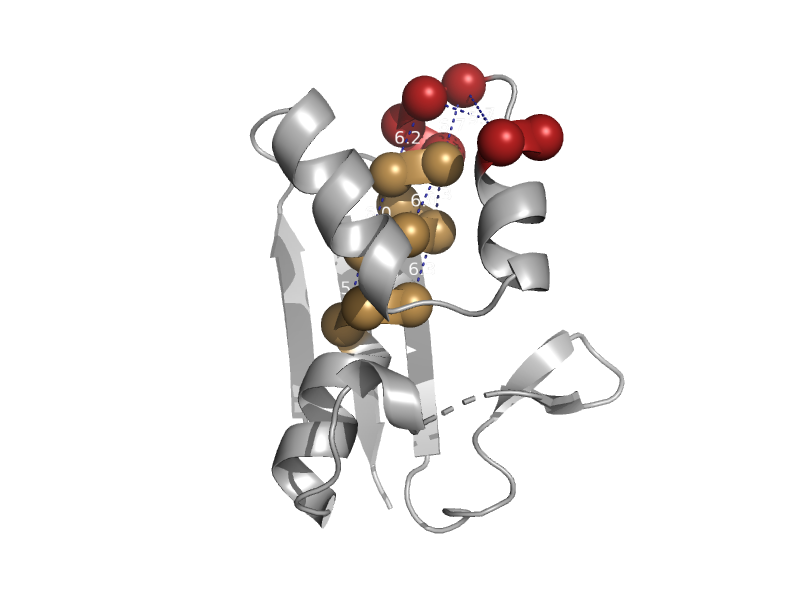 | view | |
| automated matches | 4'-Phosphopantetheinyl transferase SFP | Bacillus subtilis [TaxId: 224308] | 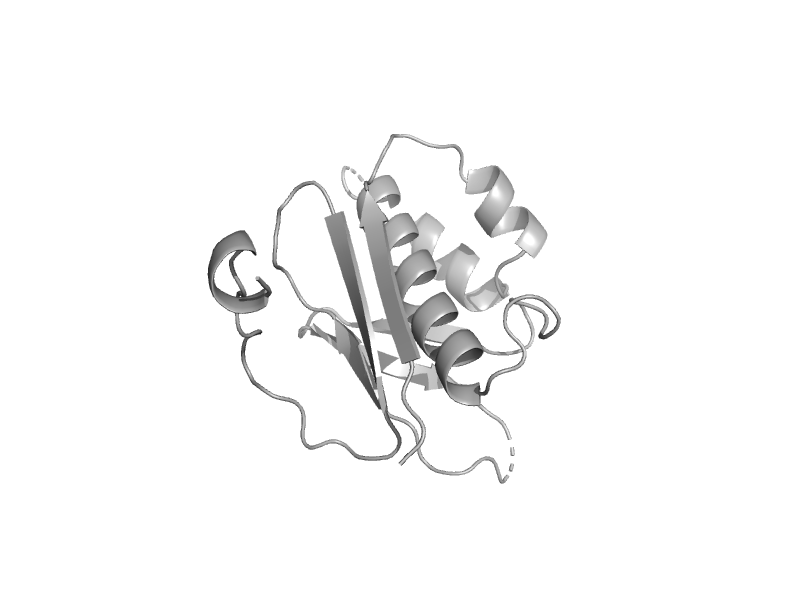 | view | |
| automated matches | automated matches | Helicobacter pylori [TaxId: 85962] | 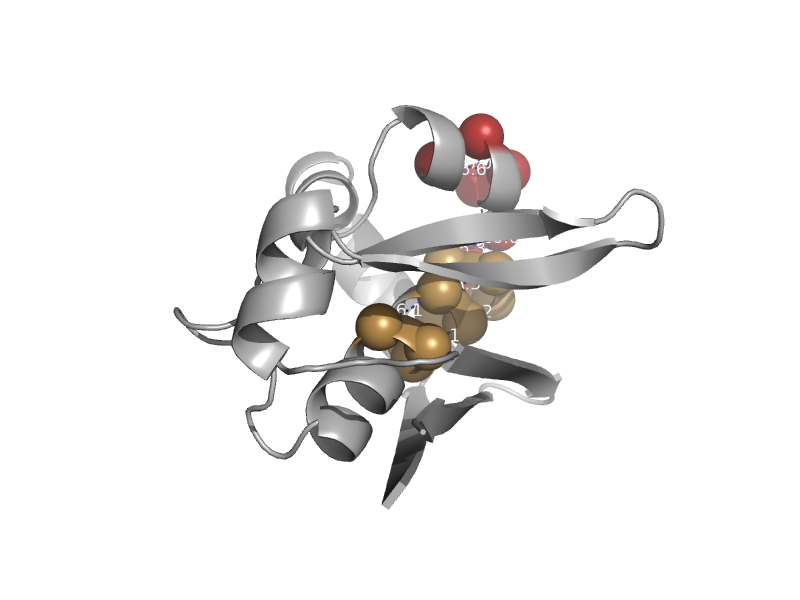 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | G | - | - | G | V | G | V | D | V | E | L | I | T | S | I | N | V | E | N | - | - | - | - | D | T | F | I | E | R | N | F | T | P | Q | E | I | E | Y | C | S | A | Q | P | - | - | - | S | V | Q | S | S | F | A | G | T | W | S | A | K | E | A | V | F | K | S | L | G | - | - | - | - | - | - | - | - | V | A | L | K | D | I | E | I | V | R | V | N | K | N | A | P | A | V | E | L | H | G | N | A | K | K | A | A | E | E | A | G | V | - | D | V | K | V | S | I | S | H | - | D | D | L | Q | A | V | A | V | A | V | S | T | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M | S | I | I | G | V | G | I | D | V | A | E | V | E | R | F | G | A | A | L | E | R | T | - | P | A | L | A | G | R | L | F | L | E | S | E | L | L | L | P | G | G | E | R | - | - | - | R | G | V | A | S | L | A | A | R | F | A | A | K | E | A | L | A | K | A | L | G | A | P | - | - | A | - | - | G | L | L | W | T | D | A | E | V | W | V | E | A | G | G | R | P | R | L | R | V | T | G | T | V | A | A | R | A | A | E | L | G | V | A | S | W | H | V | S | L | S | A | - | D | A | G | I | A | S | A | V | V | I | A | E | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | G | I | V | G | V | G | I | D | L | V | S | I | P | D | F | A | E | Q | V | - | - | - | - | D | - | - | - | - | - | T | F | T | P | G | E | R | R | D | A | S | - | - | - | - | - | - | S | A | A | R | H | L | A | A | R | W | A | A | K | E | A | V | I | K | A | W | S | G | S | R | F | A | Q | R | P | M | I | H | R | D | I | E | V | V | T | D | M | W | G | R | P | R | V | R | L | T | G | A | I | A | E | Y | L | A | - | - | D | V | - | T | I | H | V | S | L | T | H | - | E | G | D | T | A | A | A | V | A | I | L | E | A | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | I | V | G | L | G | T | D | I | A | E | I | E | R | V | E | K | A | L | A | R | S | G | E | N | F | A | R | R | I | L | T | D | S | E | L | E | Q | F | H | A | S | K | - | - | - | Q | Q | G | R | F | L | A | K | R | F | A | A | K | E | A | A | S | K | A | L | G | T | G | I | A | Q | - | - | G | V | T | F | H | D | F | T | I | S | H | D | K | L | G | K | P | L | L | I | L | S | G | Q | A | A | E | L | A | S | Q | L | Q | V | E | N | I | H | L | S | I | S | D | - | E | R | H | Y | A | - | A | T | V | I | L | E | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | M | K | I | Y | G | I | Y | M | D | R | P | L | S | - | - | - | - | Q | E | E | N | E | R | F | M | T | F | I | S | P | E | K | R | E | K | C | R | R | F | Y | H | K | E | D | A | H | R | T | L | L | G | D | V | L | V | R | S | V | I | S | R | Q | Y | Q | - | - | - | - | - | - | - | - | L | D | K | S | D | I | F | S | T | - | Q | E | Y | G | K | P | C | I | P | D | - | - | - | - | L | P | D | A | - | - | - | - | - | - | H | F | N | I | S | H | S | - | - | - | G | R | W | V | I | G | A | F | D | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | Q | P | I | G | I | D | I | E | K | T | K | P | I | S | - | - | - | - | - | - | - | L | E | I | A | K | R | F | F | S | K | T | E | Y | S | D | L | L | A | K | D | D | - | - | E | Q | T | D | Y | F | Y | H | L | W | S | M | K | E | S | F | I | K | Q | E | G | K | - | - | - | - | G | L | S | L | P | L | D | S | F | S | V | R | L | H | Q | D | G | Q | V | S | I | E | L | - | - | - | - | P | S | H | S | - | - | - | - | - | - | P | C | Y | I | K | T | Y | E | V | D | P | G | Y | K | M | A | V | C | A | A | H | P | D | F | P | E | D | I | T | M | V | S | Y | E | E | L | L |
| - | - | - | - | M | I | G | I | D | I | V | S | I | A | R | I | E | K | C | V | K | R | F | K | M | K | F | L | E | R | F | L | S | P | S | E | I | V | L | C | K | D | - | - | - | - | - | - | K | S | S | S | I | A | G | F | F | A | L | K | E | A | C | S | K | A | L | Q | V | G | I | G | K | - | - | E | L | S | F | L | D | I | K | I | S | K | S | P | K | N | A | P | L | I | T | L | S | - | - | - | K | E | K | M | D | Y | F | N | I | Q | S | L | S | A | S | I | S | H | - | D | A | G | F | A | I | A | V | V | V | V | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|
