solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1hska2 ( 209 ) gkmteiqakmddlterreskqPl--eypscgsVFqrpp------------
d1uxya2 ( 201 ) vtpqqvfnavchmrttklpdpkvngnagaFFknpvvsaetAkaLlsq
aaaaaaaaaaaaaaa bb
60 70 80 90 100
d1hska2 ( 245 ) --------------ghfagkLIqdSnLqghrigGVeVStkhAgfMVnvdn
d1uxya2 ( 248 ) fptApnypqadgsvkLaAgwLIdqcqLkGmqiggAAVhrqqAlVLinedn
aaaaaaa bb bbb bbb
110 120 130 140
d1hska2 ( 281 ) GtatdyenLIhyVqktVkekfgieLnrevriig--------ehpk
d1uxya2 ( 298 ) AksedvvqLAhhVrqkVGekFnVwLePevrfigasgevsavetIs
aaaaaaaaaaaaaaaaaaa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain | Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain | Staphylococcus aureus [TaxId: 1280] | 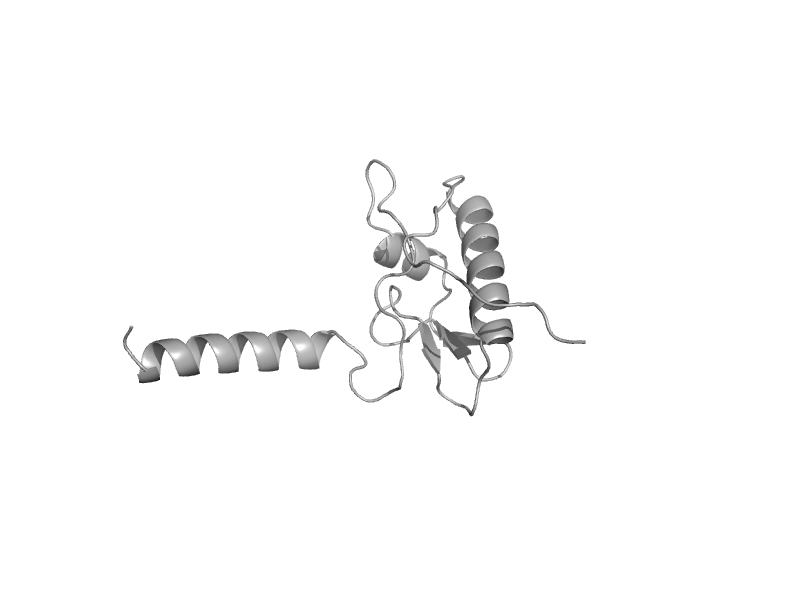 | view | |
| Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain | Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain | Escherichia coli [TaxId: 562] | 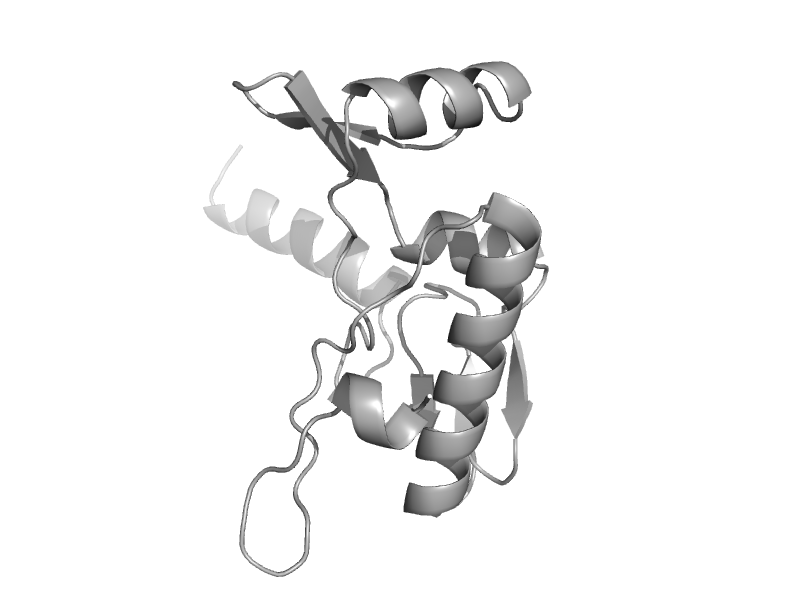 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | K | M | T | E | I | Q | A | K | M | D | D | L | T | E | R | R | E | S | K | Q | P | L | - | - | E | Y | P | S | C | G | S | V | F | Q | R | P | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | H | F | A | G | K | L | I | Q | D | S | N | L | Q | G | H | R | I | G | G | V | E | V | S | T | K | H | A | G | F | M | V | N | V | D | N | G | T | A | T | D | Y | E | N | L | I | H | Y | V | Q | K | T | V | K | E | K | F | G | I | E | L | N | R | E | V | R | I | I | G | - | - | - | - | - | - | - | - | E | H | P | K |
| - | - | - | V | T | P | Q | Q | V | F | N | A | V | C | H | M | R | T | T | K | L | P | D | P | K | V | N | G | N | A | G | A | F | F | K | N | P | V | V | S | A | E | T | A | K | A | L | L | S | Q | F | P | T | A | P | N | Y | P | Q | A | D | G | S | V | K | L | A | A | G | W | L | I | D | Q | C | Q | L | K | G | M | Q | I | G | G | A | A | V | H | R | Q | Q | A | L | V | L | I | N | E | D | N | A | K | S | E | D | V | V | Q | L | A | H | H | V | R | Q | K | V | G | E | K | F | N | V | W | L | E | P | E | V | R | F | I | G | A | S | G | E | V | S | A | V | E | T | I | S |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1hska2 | A | P61431 | GO:0005737 |
| d1hska2 | A | P61431 | GO:0005829 |
| d1hska2 | A | P61431 | GO:0008360 |
| d1hska2 | A | P61431 | GO:0008762 |
| d1hska2 | A | P61431 | GO:0009252 |
| d1hska2 | A | P61431 | GO:0016491 |
| d1hska2 | A | P61431 | GO:0050660 |
| d1hska2 | A | P61431 | GO:0051301 |
| d1hska2 | A | P61431 | GO:0071555 |
| d1hska2 | A | P61431 | GO:0071949 |
| d1uxya2 | A | P08373 | GO:0005737 |
| d1uxya2 | A | P08373 | GO:0005829 |
| d1uxya2 | A | P08373 | GO:0008360 |
| d1uxya2 | A | P08373 | GO:0008762 |
| d1uxya2 | A | P08373 | GO:0009252 |
| d1uxya2 | A | P08373 | GO:0016491 |
| d1uxya2 | A | P08373 | GO:0050660 |
| d1uxya2 | A | P08373 | GO:0051301 |
| d1uxya2 | A | P08373 | GO:0071555 |
| d1uxya2 | A | P08373 | GO:0071949 |
