solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1hp1a1 ( 363 ) kigeTngrLeGdrdkVrfvqtnMGrLILaAQmdrT-----gAdFAVM
d2z1aa1 ( 330 ) mqqvIaeAkvdLvGeravVrrreSnLGNLITdGMlwkTr--nagTqIALQ
d3c9fa1 ( 338 ) dtligyVktnyyvDy-vpidhpkSIFnLLalkILktLpkskheeRITII
bbb aaaaaaa bbbb
60 70 80 90 100
d1hp1a1 ( 405 ) SGggIrdSIeagdIsyknVlkVQpfgnvVVyadMtGkeVidYLtaVAqmk
d2z1aa1 ( 378 ) nGggIrasIpkgpItvgkVyeVLpfgntLVVMdLkGkeIlaALenGVsqW
d3c9fa1 ( 386 ) nTgSIrydLykgpYtidskfiVSpfeniWvnItVpKsvAtkVAakLndyi
b333 bbb aaaaaaa bbbbbbbaaaa aaaaa
110 120 130 140 150
d1hp1a1 ( 455 ) p-dsgaYPQFA---nVsFvak-----d----------------------g
d2z1aa1 ( 428 ) entagrFLQVS---gLrYAFdLsrpag----------------------s
d3c9fa1 ( 438 ) sa----srLkPphQydlqvqqeklpkgyvthddfgadgddtlhravvnfp
160 170 180 190 200
d1hp1a1 ( 474 ) kLndLkIk----gepVdpaktyrMATLnfNAtggdgYprldnk---pgyv
d2z1aa1 ( 453 ) RVvrVeVktekgyvpLdleatyrVVVNnfIAnGgdgFtvLkeA--qgyrv
d3c9fa1 ( 497 ) VpnVIqSveindevd----spVnLVFYsfItpnIiwALkelnfsteqvpt
bbbb bbbbbbb a b
210 220 230
d1hp1a1 ( 517 ) nt-gfiDAeVLkayIqksspLdvsvyepkgeVswq
d2z1aa1 ( 501 ) dt-gfsdAeSFmdyLkelkvVea---glegrIeVlnep
d3c9fa1 ( 543 ) pysdiyLGtLLnefVan--------------------n
bb aaaaaaaaaa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| 5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain | 5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain | Escherichia coli [TaxId: 562] | 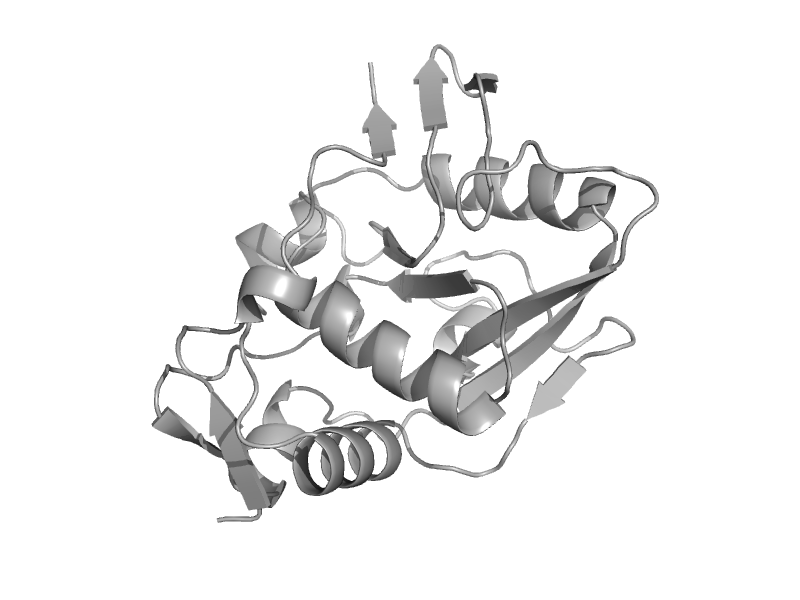 | view | |
| 5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain | 5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain | Thermus thermophilus [TaxId: 274] | 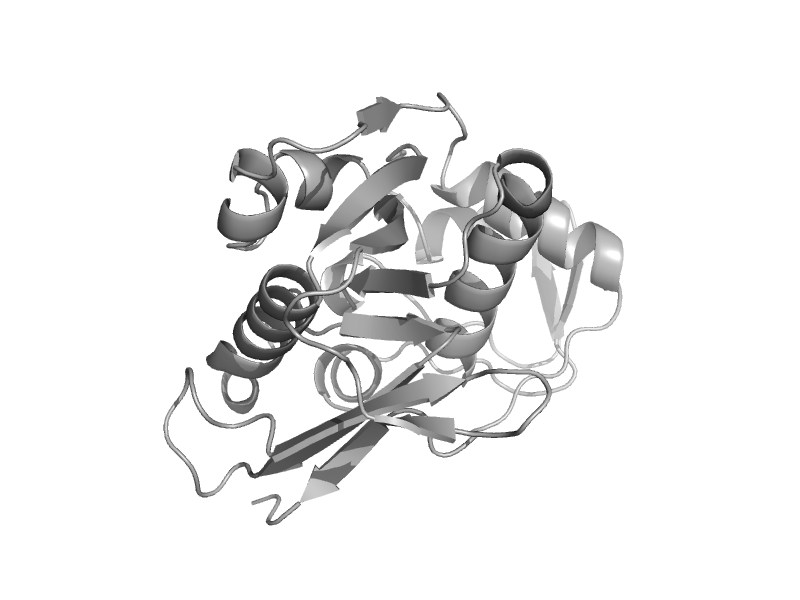 | view | |
| 5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain | 5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain | Yeast (Candida albicans) [TaxId: 5476] | 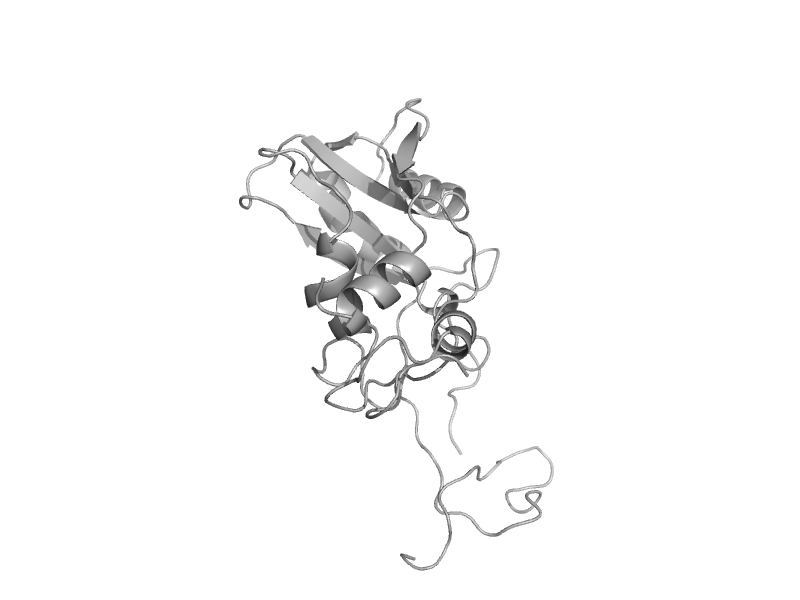 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | K | I | G | E | T | N | G | R | L | E | G | D | R | D | K | V | R | F | V | Q | T | N | M | G | R | L | I | L | A | A | Q | M | D | R | T | - | - | - | - | - | G | A | D | F | A | V | M | S | G | G | G | I | R | D | S | I | E | A | G | D | I | S | Y | K | N | V | L | K | V | Q | P | F | G | N | V | V | V | Y | A | D | M | T | G | K | E | V | I | D | Y | L | T | A | V | A | Q | M | K | P | - | D | S | G | A | Y | P | Q | F | A | - | - | - | N | V | S | F | V | A | K | - | - | - | - | - | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | K | L | N | D | L | K | I | K | - | - | - | - | G | E | P | V | D | P | A | K | T | Y | R | M | A | T | L | N | F | N | A | T | G | G | D | G | Y | P | R | L | D | N | K | - | - | - | P | G | Y | V | N | T | - | G | F | I | D | A | E | V | L | K | A | Y | I | Q | K | S | S | P | L | D | V | S | V | Y | E | P | K | G | E | V | S | W | Q | - | - | - |
| M | Q | Q | V | I | A | E | A | K | V | D | L | V | G | E | R | A | V | V | R | R | R | E | S | N | L | G | N | L | I | T | D | G | M | L | W | K | T | R | - | - | N | A | G | T | Q | I | A | L | Q | N | G | G | G | I | R | A | S | I | P | K | G | P | I | T | V | G | K | V | Y | E | V | L | P | F | G | N | T | L | V | V | M | D | L | K | G | K | E | I | L | A | A | L | E | N | G | V | S | Q | W | E | N | T | A | G | R | F | L | Q | V | S | - | - | - | G | L | R | Y | A | F | D | L | S | R | P | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | R | V | V | R | V | E | V | K | T | E | K | G | Y | V | P | L | D | L | E | A | T | Y | R | V | V | V | N | N | F | I | A | N | G | G | D | G | F | T | V | L | K | E | A | - | - | Q | G | Y | R | V | D | T | - | G | F | S | D | A | E | S | F | M | D | Y | L | K | E | L | K | V | V | E | A | - | - | - | G | L | E | G | R | I | E | V | L | N | E | P |
| - | D | T | L | I | G | Y | V | K | T | N | Y | Y | V | D | Y | - | V | P | I | D | H | P | K | S | I | F | N | L | L | A | L | K | I | L | K | T | L | P | K | S | K | H | E | E | R | I | T | I | I | N | T | G | S | I | R | Y | D | L | Y | K | G | P | Y | T | I | D | S | K | F | I | V | S | P | F | E | N | I | W | V | N | I | T | V | P | K | S | V | A | T | K | V | A | A | K | L | N | D | Y | I | S | A | - | - | - | - | S | R | L | K | P | P | H | Q | Y | D | L | Q | V | Q | Q | E | K | L | P | K | G | Y | V | T | H | D | D | F | G | A | D | G | D | D | T | L | H | R | A | V | V | N | F | P | V | P | N | V | I | Q | S | V | E | I | N | D | E | V | D | - | - | - | - | S | P | V | N | L | V | F | Y | S | F | I | T | P | N | I | I | W | A | L | K | E | L | N | F | S | T | E | Q | V | P | T | P | Y | S | D | I | Y | L | G | T | L | L | N | E | F | V | A | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N |
