solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d2pbdp- ( 1 ) agwnayid-nLmadg----tCqDAAIVGykdspsvwaAvpgktFvnItpa
d3d9ya- ( 1 ) swqayVdtsLlgtg----kIdrAAIVsr--aGdswaasa--gF-nLspq
d4qwoa1 ( 2 ) aewhkIie-dIsknn----kFedAAIVdykttknvlAaipnrtFakInpg
d5em0a1 ( 2 ) swqtyVddhLmdiegtgqhLtSAAIFGt--dgtvwAksa--sFpefkpn
aaaaaa a bbbbbb bbbb aa
60 70 80 90 100
d2pbdp- ( 46 ) evgvLvg--kdrssFyvnGltLGgqkCsVirdsLlqdgefsMdLrTkstg
d3d9ya- ( 42 ) eiqgLaaGFqdppsmfgtgIiLagqkY-TirAe-g----rSIyGklq---
d4qwoa1 ( 47 ) eVipLit---nhnil---kPlIGqkfCivytnsld-entyA-ELltgy--
d5em0a1 ( 48 ) EIdaIikeFneagqLaptgLfLGgakYmviqgeag----aVirGkkg---
aaaaaaa bbb bbbbbbbbb bbbbbb
110 120 130 140
d2pbdp- ( 94 ) gaptfnVTVTkTdkTLVLLMGkegvhgglInkkCyemAshLrrsqy
d3d9ya- ( 84 ) ---keGIICVATklCILVShYpetTlpgeAakiTealAdyLvgvgy
d4qwoa1 ( 89 ) -apvspIVIArThtALIFL-GkpttsrrdVyrtCrdhAtrvratgn
d5em0a1 ( 91 ) ---aGGIÇIkkTgqAMVFGiYdepVapgqÇnmvVerLGdyLldqgm
bbbbbb bbbbbbb aaaaaaaaaaaaaaaaa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| automated matches | Profilin (actin-binding protein) | Human (Homo sapiens) [TaxId: 9606] | 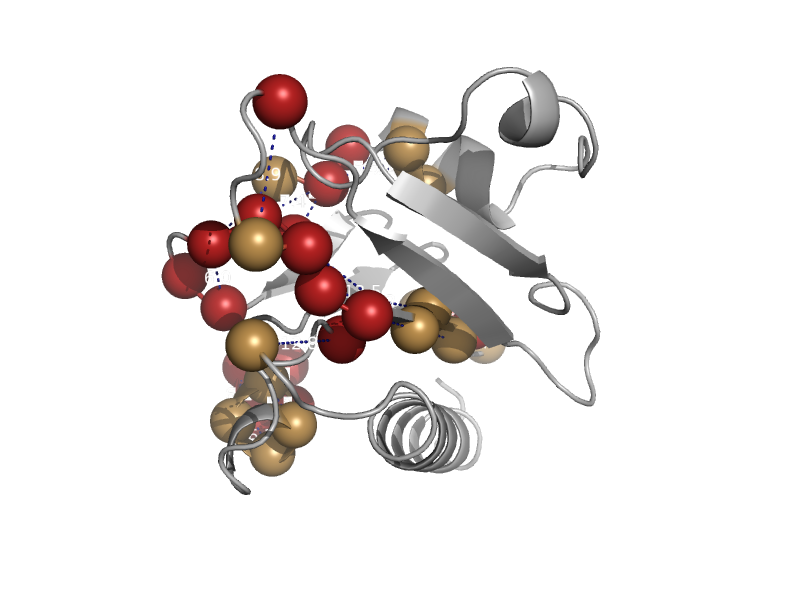 | view | |
| automated matches | automated matches | Fission yeast (Schizosaccharomyces pombe) [TaxId: 4896] | 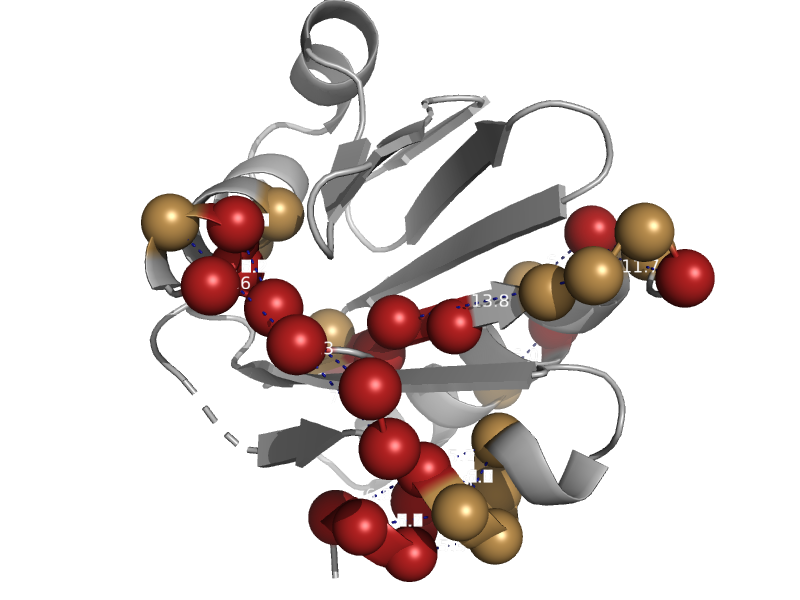 | view | |
| automated matches | automated matches | Monkeypox virus [TaxId: 619591] | 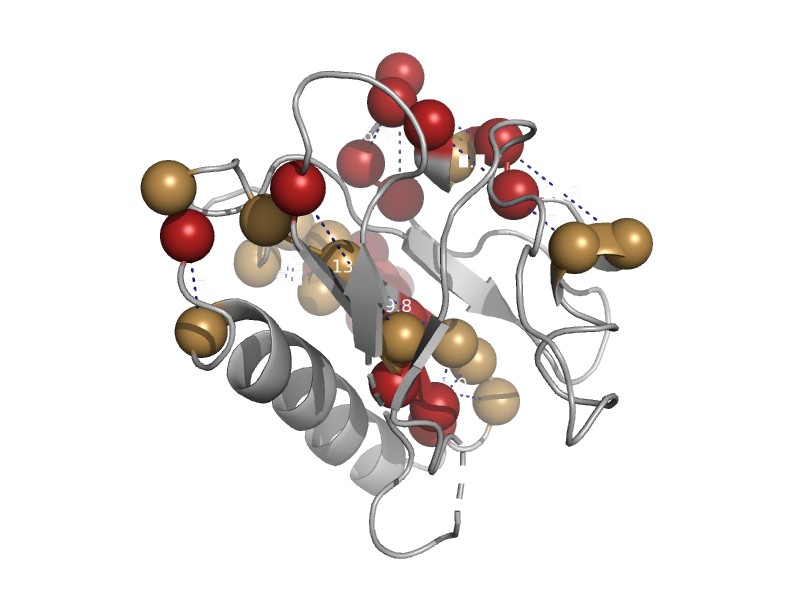 | view | |
| automated matches | Profilin (actin-binding protein) | Artemisia vulgaris [TaxId: 4220] | 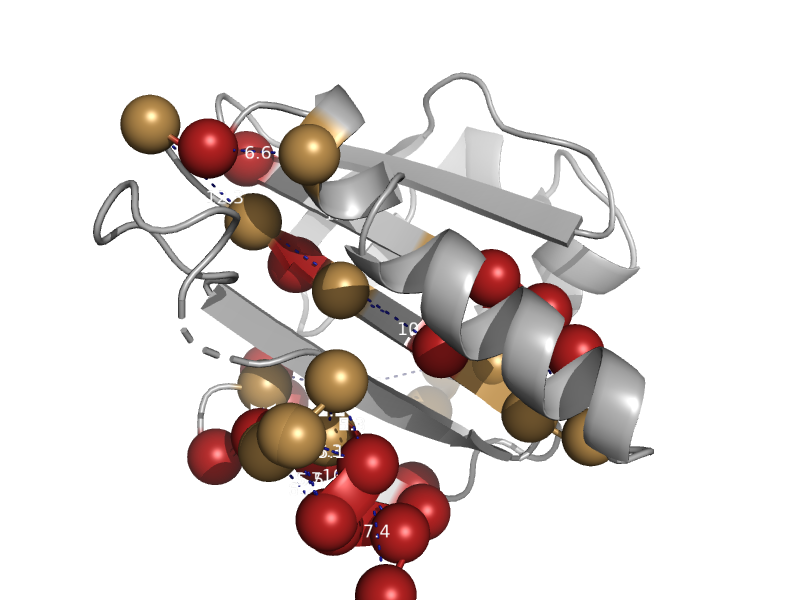 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | W | N | A | Y | I | D | - | N | L | M | A | D | G | - | - | - | - | T | C | Q | D | A | A | I | V | G | Y | K | D | S | P | S | V | W | A | A | V | P | G | K | T | F | V | N | I | T | P | A | E | V | G | V | L | V | G | - | - | K | D | R | S | S | F | Y | V | N | G | L | T | L | G | G | Q | K | C | S | V | I | R | D | S | L | L | Q | D | G | E | F | S | M | D | L | R | T | K | S | T | G | G | A | P | T | F | N | V | T | V | T | K | T | D | K | T | L | V | L | L | M | G | K | E | G | V | H | G | G | L | I | N | K | K | C | Y | E | M | A | S | H | L | R | R | S | Q | Y |
| - | S | W | Q | A | Y | V | D | T | S | L | L | G | T | G | - | - | - | - | K | I | D | R | A | A | I | V | S | R | - | - | A | G | D | S | W | A | A | S | A | - | - | G | F | - | N | L | S | P | Q | E | I | Q | G | L | A | A | G | F | Q | D | P | P | S | M | F | G | T | G | I | I | L | A | G | Q | K | Y | - | T | I | R | A | E | - | G | - | - | - | - | R | S | I | Y | G | K | L | Q | - | - | - | - | - | - | K | E | G | I | I | C | V | A | T | K | L | C | I | L | V | S | H | Y | P | E | T | T | L | P | G | E | A | A | K | I | T | E | A | L | A | D | Y | L | V | G | V | G | Y |
| A | E | W | H | K | I | I | E | - | D | I | S | K | N | N | - | - | - | - | K | F | E | D | A | A | I | V | D | Y | K | T | T | K | N | V | L | A | A | I | P | N | R | T | F | A | K | I | N | P | G | E | V | I | P | L | I | T | - | - | - | N | H | N | I | L | - | - | - | K | P | L | I | G | Q | K | F | C | I | V | Y | T | N | S | L | D | - | E | N | T | Y | A | - | E | L | L | T | G | Y | - | - | - | A | P | V | S | P | I | V | I | A | R | T | H | T | A | L | I | F | L | - | G | K | P | T | T | S | R | R | D | V | Y | R | T | C | R | D | H | A | T | R | V | R | A | T | G | N |
| - | S | W | Q | T | Y | V | D | D | H | L | M | D | I | E | G | T | G | Q | H | L | T | S | A | A | I | F | G | T | - | - | D | G | T | V | W | A | K | S | A | - | - | S | F | P | E | F | K | P | N | E | I | D | A | I | I | K | E | F | N | E | A | G | Q | L | A | P | T | G | L | F | L | G | G | A | K | Y | M | V | I | Q | G | E | A | G | - | - | - | - | A | V | I | R | G | K | K | G | - | - | - | - | - | - | A | G | G | I | C | I | K | K | T | G | Q | A | M | V | F | G | I | Y | D | E | P | V | A | P | G | Q | C | N | M | V | V | E | R | L | G | D | Y | L | L | D | Q | G | M |
| Member | Chain | UniProtKB | GO term |
|---|
