solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1qtsa2 ( 825 ) fFqptemasqdFfqrwkqlsnpqqevqnifkAkhpMdteiTkakIigf-g
d1r4xa2 ( 763 ) hiqkvmkl--nFeaawdev-gdefekeetftlstiktLeeAVgnivkfLg
d2g30a2 ( 825 ) lfvdGkMerqvFlatwkdI-pnenelqfqIk-eChlnAdtVsskLqnn-n
aaaaa bbbbb aaaaaaaaaa
60 70 80 90 100
d1qtsa2 ( 874 ) SAlLeeVdpn----p-anfVGAGiihtkttqIgClLrLepnl-qaqmYrL
d1r4xa2 ( 810 ) MhpCersdkVpdnknthtLlLAGvFr---gghdILVrSrLllldtVtMqV
d2g30a2 ( 873 ) VytIakrnve----gqdmLyQSLkl--tn-giwILAeLriqp-gnpnYtL
bb bbbbbb bbbbbbbb bbb
110 120
d1qtsa2 ( 918 ) TLrTskdtvSqrlCelLseqF
d1r4xa2 ( 857 ) TArSleelPVdiIlasv
d2g30a2 ( 915 ) SLKCrapevSqyIyqvYdsiLkn
bbbb aaaaaaa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Alpa-adaptin AP2, C-terminal subdomain | Clathrin adaptor appendage, alpha and beta chain-specific domain | Mouse (Mus musculus) [TaxId: 10090] | 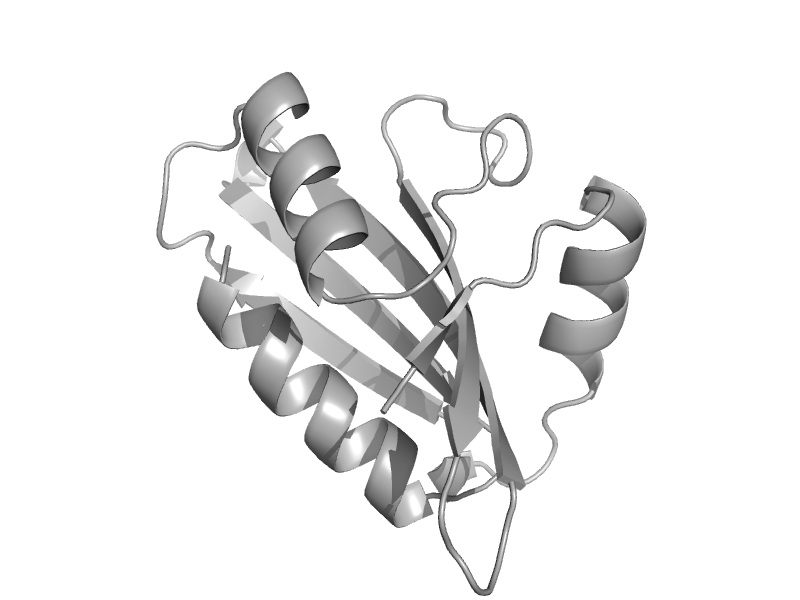 | view | |
| Coatomer gamma subunit, C-terminal subdomain | Coatomer appendage domain | Human (Homo sapiens) [TaxId: 9606] | 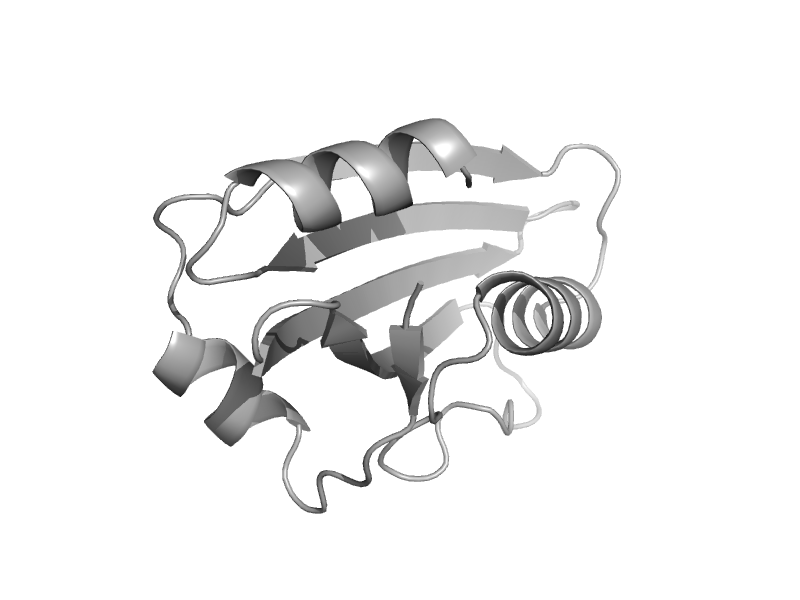 | view | |
| Beta2-adaptin AP2, C-terminal subdomain | Clathrin adaptor appendage, alpha and beta chain-specific domain | Human (Homo sapiens) [TaxId: 9606] | 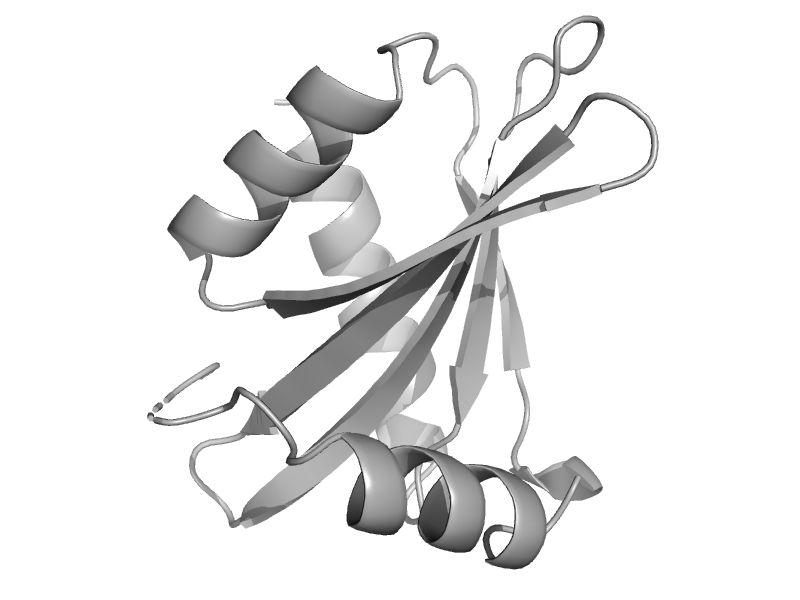 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F | F | Q | P | T | E | M | A | S | Q | D | F | F | Q | R | W | K | Q | L | S | N | P | Q | Q | E | V | Q | N | I | F | K | A | K | H | P | M | D | T | E | I | T | K | A | K | I | I | G | F | - | G | S | A | L | L | E | E | V | D | P | N | - | - | - | - | P | - | A | N | F | V | G | A | G | I | I | H | T | K | T | T | Q | I | G | C | L | L | R | L | E | P | N | L | - | Q | A | Q | M | Y | R | L | T | L | R | T | S | K | D | T | V | S | Q | R | L | C | E | L | L | S | E | Q | F | - | - |
| H | I | Q | K | V | M | K | L | - | - | N | F | E | A | A | W | D | E | V | - | G | D | E | F | E | K | E | E | T | F | T | L | S | T | I | K | T | L | E | E | A | V | G | N | I | V | K | F | L | G | M | H | P | C | E | R | S | D | K | V | P | D | N | K | N | T | H | T | L | L | L | A | G | V | F | R | - | - | - | G | G | H | D | I | L | V | R | S | R | L | L | L | L | D | T | V | T | M | Q | V | T | A | R | S | L | E | E | L | P | V | D | I | I | L | A | S | V | - | - | - | - | - | - |
| L | F | V | D | G | K | M | E | R | Q | V | F | L | A | T | W | K | D | I | - | P | N | E | N | E | L | Q | F | Q | I | K | - | E | C | H | L | N | A | D | T | V | S | S | K | L | Q | N | N | - | N | V | Y | T | I | A | K | R | N | V | E | - | - | - | - | G | Q | D | M | L | Y | Q | S | L | K | L | - | - | T | N | - | G | I | W | I | L | A | E | L | R | I | Q | P | - | G | N | P | N | Y | T | L | S | L | K | C | R | A | P | E | V | S | Q | Y | I | Y | Q | V | Y | D | S | I | L | K | N |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1qtsa2 | A | P17427 | GO:0006886 |
| d1qtsa2 | A | P17427 | GO:0016192 |
| d1qtsa2 | A | P17427 | GO:0030117 |
| d1qtsa2 | A | P17427 | GO:0030131 |
| d1qtsa2 | A | P17427 | GO:0006886 |
| d1qtsa2 | A | P17427 | GO:0016192 |
| d1qtsa2 | A | P17427 | GO:0030117 |
| d1qtsa2 | A | P17427 | GO:0030131 |
| d1r4xa2 | A | Q9Y678 | GO:0005198 |
| d1r4xa2 | A | Q9Y678 | GO:0006886 |
| d1r4xa2 | A | Q9Y678 | GO:0016192 |
| d1r4xa2 | A | Q9Y678 | GO:0030117 |
| d1r4xa2 | A | Q9Y678 | GO:0030126 |
| d1r4xa2 | A | Q9Y678 | GO:0005198 |
| d1r4xa2 | A | Q9Y678 | GO:0006886 |
| d1r4xa2 | A | Q9Y678 | GO:0016192 |
| d1r4xa2 | A | Q9Y678 | GO:0030117 |
| d1r4xa2 | A | Q9Y678 | GO:0030126 |
| d2g30a2 | A | P63010 | GO:0006886 |
| d2g30a2 | A | P63010 | GO:0016192 |
| d2g30a2 | A | P63010 | GO:0030117 |
| d2g30a2 | A | P63010 | GO:0030131 |
| d2g30a2 | A | Q5SW96 | GO:0006886 |
| d2g30a2 | A | Q5SW96 | GO:0016192 |
| d2g30a2 | A | Q5SW96 | GO:0030117 |
| d2g30a2 | A | Q5SW96 | GO:0030131 |
| d2g30a2 | A | P63010 | GO:0006886 |
| d2g30a2 | A | P63010 | GO:0016192 |
| d2g30a2 | A | P63010 | GO:0030117 |
| d2g30a2 | A | P63010 | GO:0030131 |
| d2g30a2 | A | Q5SW96 | GO:0006886 |
| d2g30a2 | A | Q5SW96 | GO:0016192 |
| d2g30a2 | A | Q5SW96 | GO:0030117 |
| d2g30a2 | A | Q5SW96 | GO:0030131 |
