solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1f08a1 ( 159 )
d1m55a- ( 1 ) matFYEViVrVPf---------dveehLpgIs------------------
d1p4da- ( 1 ) mMsIaqVr--sagsAGnyYtdkdnyyVLgsmgerWagrGAeqLglqgs
d1r9wa- ( 210 )
d3l57a- ( 1 ) mLdIttItrqnVtsVvgyysdaddyys--ssfTsWqgtGAealglsgd
d4nbpa- ( 135 )
d6q1ma- ( 7 )
60 70 80 90 100
d1f08a1 ( 159 ) atvfklglFksLFlcsFhd--ITrlfkndkttnqqWVLAVfgLa------
d1m55a- ( 24 ) ---dsFvdwvt-------gqiwelpp-------------------eSdLn
d1p4da- ( 47 ) VdkdvFtrLLeGrLp--dgadlSrmqdgsnrHrpGydLtFsApkSVSMMA
d1r9wa- ( 210 ) kqgamlavFkdtYglsFtd--LVr-------tctdWVTAIFgVn------
d3l57a- ( 50 ) VesarFkeLLvGeid--tfthMqR---gakkerLGYdLtFsApkgVSmqA
d4nbpa- ( 135 ) dpdFpvdLhaf--Lsqav--f---------sntvasFAVYTt-------
d6q1ma- ( 7 ) frvyskyLfLtYpqCtl-----
aaaaaaa bbbbbbb
110 120 130 140 150
d1f08a1 ( 201 ) ------------evffeaSfelLkkq--------------C-sFLQMqkr
d1m55a- ( 45 ) ltlVeqpqLTVAdrIrrvFLyeWnkfSk-----------qeSkfFVQFEk
d1p4da- ( 95 ) MLGgDkrLidAHnqAVdfAVrqVEalASTrvmtdgqsetvlTgnLVMALF
d1r9wa- ( 252 ) ------------ptiAegFktlIqpf--------------I-lYAHIQcl
d3l57a- ( 98 ) lihgdktIieaHekAVaaAVreAeklAqArtg----svtqnTnnLVVATF
d4nbpa- ( 166 ) ------------kekAqiLykkLmek--------------y-sVtFISrH
d6q1ma- ( 24 ) ----------ePqyALdsLrtlLnky--------------eplYIAAVre
aaaaaaaaaaaa bbbbb
160 170 180 190 200
d1f08a1 ( 224 ) she-----ggtCAVYLIcFntaksRetVrnlManmLnv--reecLMLqPP
d1m55a- ( 84 ) gs-------eyfhLHTLVets---------------gIs-----------
d1p4da- ( 145 ) NHDtsrdqePqLhThAVVANVTqhn-----------geWktL-ssdkvgk
d1r9wa- ( 275 ) dck-----wgvLILALLrYkcgksrltVAkgLstlLhv--petcMLIqpP
d3l57a- ( 148 ) Rhet---rdPdLhThAFVMNMTqred----------gqwraL-k------
d4nbpa- ( 189 ) gfg-----g--hNILFFlTphrhrvsaInnyCqklCt----fsfLiCkgV
d6q1ma- ( 50 ) lhe----dgsphLHVLVQNlrAsI--tnpnaLn--lrmdtspfsifhPnI
bb bbbbbbbbb bb
210 220 230 240 250
d1f08a1 ( 267 ) --ki-----rglsAAlfWfksslsp-------------------------
d1m55a- ( 101 ) -------smvLgryVsqIraqLvkvVFq-giePqIn-dWVaitkvkkgga
d1p4da- ( 183 ) tgFienVyanqiafGrlYrekLkeqVealGYeTevvgkhgmWeMpgVpve
d1r9wa- ( 318 ) --kl-----rssvAAlyWyrtgisn-------------------------
d3l57a- ( 181 ) ---ndeLmrnkmhLgdvYkqeLaleLtkagyeLrynsn-ntFdmAh
d4nbpa- ( 228 ) --n----------keylFYsaLcrq-------------------------
d6q1ma- ( 93 ) qaAk-----dCnqvrdyItkeVdsdv------------------------
aaaaaaaaaa
260 270 280 290 300
d1f08a1 ( 285 ) -------------------aTlkhg-----alpewIraqttln
d1m55a- ( 142 ) nkvvdsgyIpayLLpkvQpeLQWAwTnldeYklAalnleeRkrLvaqfla
d1p4da- ( 233 ) aFsvdpeikmaeWmqt-lketgfd--------irayrdaAdqradlrtlt
d1r9wa- ( 336 ) -------------------iSevmg-----dtPewIqrlTiiq
d3l57a-
d4nbpa- ( 241 ) -------------------pYavveesiqgGLkehdfnpe
d6q1ma- ( 114 ) -------------------ntaewgtfvavs
bbbb
d1f08a1
d1m55a- ( 192 ) es
d1p4da- ( 306 ) pg
d1r9wa-
d3l57a-
d4nbpa-
d6q1ma-
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Replication initiation protein E1 | Replication initiation protein E1 | Bovine papillomavirus [TaxId: 10571] | 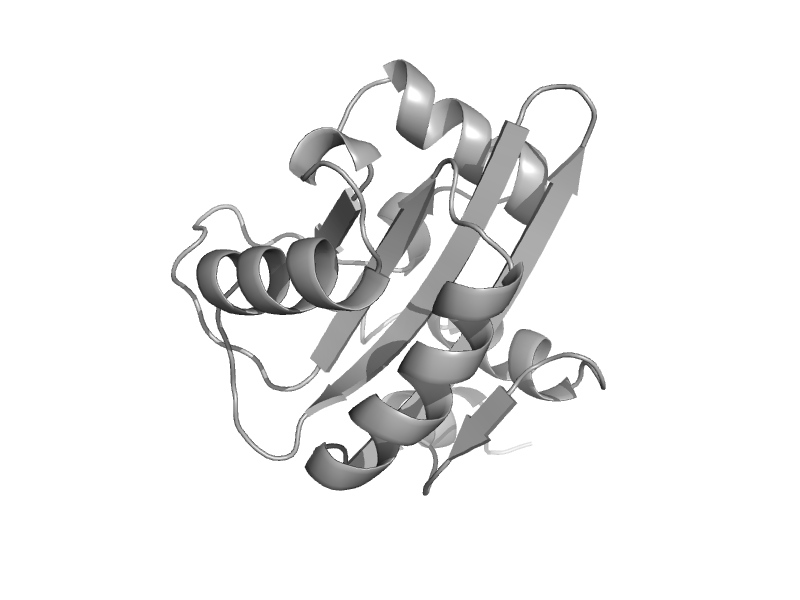 | view | |
| Replication protein Rep, nuclease domain | Replication protein Rep, nuclease domain | Adeno-associated virus, aav-5 [TaxId: 272636] | 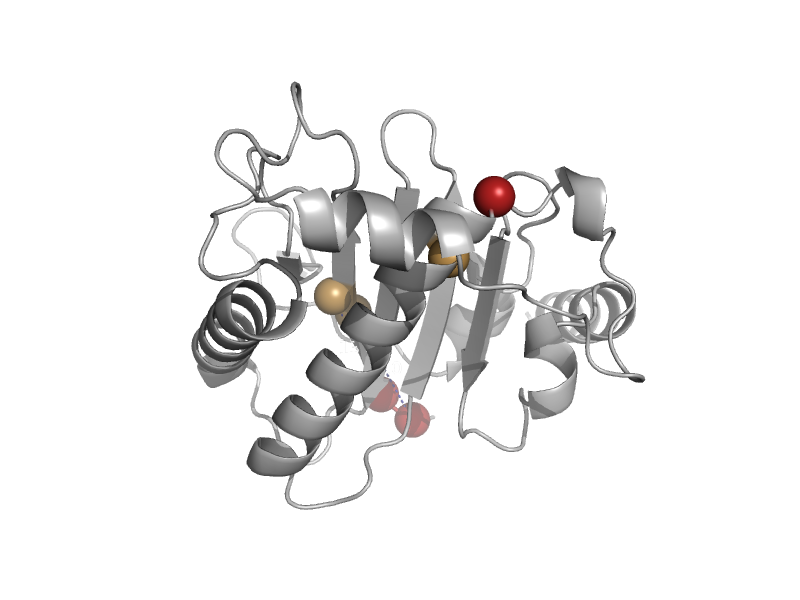 | view | |
| F factor TraI relaxase | Relaxase domain | Escherichia coli [TaxId: 562] | 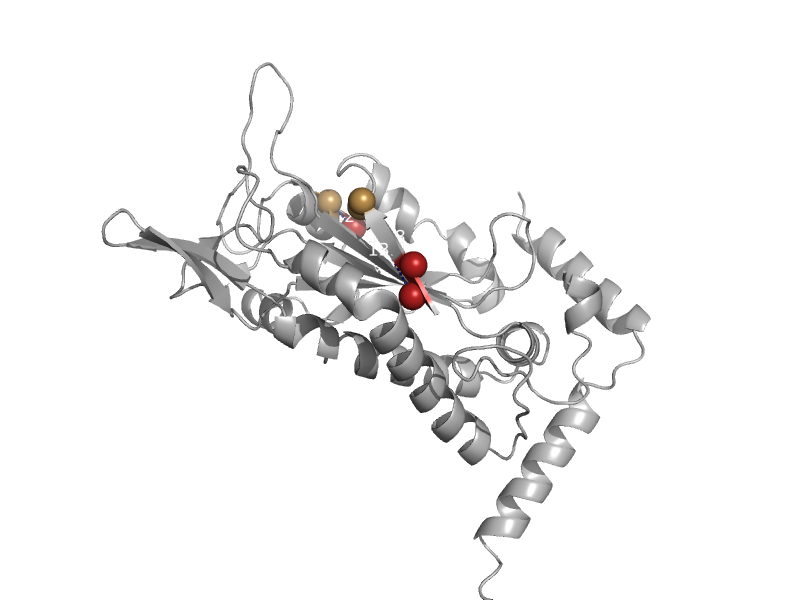 | view | |
| Replication initiation protein E1 | Replication initiation protein E1 | Human papillomavirus type 18 [TaxId: 333761] | 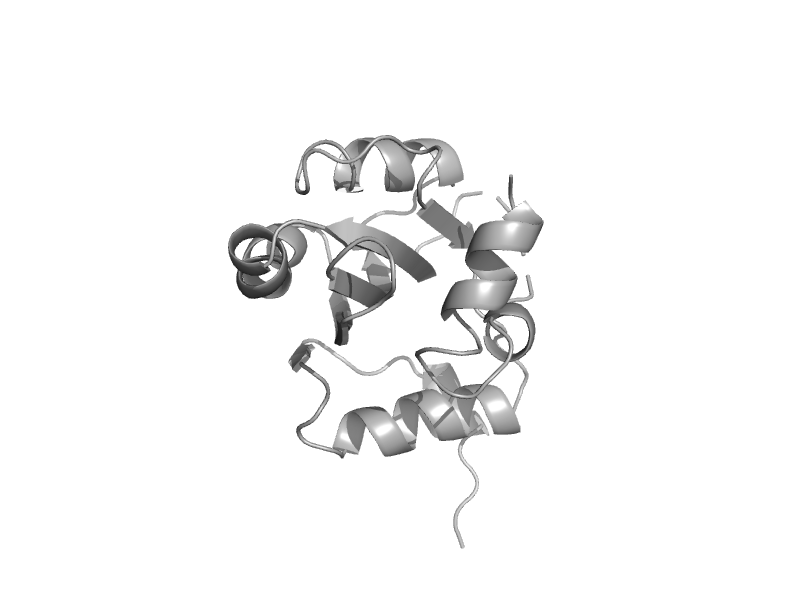 | view | |
| automated matches | automated matches | Escherichia coli [TaxId: 562] |  | view | |
| The origin DNA-binding domain of SV40 T-antigen | The origin DNA-binding domain of SV40 T-antigen | Jc polyomavirus [TaxId: 10632] |  | view | |
| automated matches | automated matches | Wheat dwarf virus [TaxId: 10834] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | T | V | F | K | L | G | L | F | K | S | L | F | L | C | S | F | H | D | - | - | I | T | R | L | F | K | N | D | K | T | T | N | Q | Q | W | V | L | A | V | F | G | L | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | V | F | F | E | A | S | F | E | L | L | K | K | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | - | S | F | L | Q | M | Q | K | R | S | H | E | - | - | - | - | - | G | G | T | C | A | V | Y | L | I | C | F | N | T | A | K | S | R | E | T | V | R | N | L | M | A | N | M | L | N | V | - | - | R | E | E | C | L | M | L | Q | P | P | - | - | K | I | - | - | - | - | - | R | G | L | S | A | A | L | F | W | F | K | S | S | L | S | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | T | L | K | H | G | - | - | - | - | - | A | L | P | E | W | I | R | A | Q | T | T | L | N | - | - | - | - | - | - | - | - | - |
| M | A | T | F | Y | E | V | I | V | R | V | P | F | - | - | - | - | - | - | - | - | - | D | V | E | E | H | L | P | G | I | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | S | F | V | D | W | V | T | - | - | - | - | - | - | - | G | Q | I | W | E | L | P | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | S | D | L | N | L | T | L | V | E | Q | P | Q | L | T | V | A | D | R | I | R | R | V | F | L | Y | E | W | N | K | F | S | K | - | - | - | - | - | - | - | - | - | - | - | Q | E | S | K | F | F | V | Q | F | E | K | G | S | - | - | - | - | - | - | - | E | Y | F | H | L | H | T | L | V | E | T | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | I | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | M | V | L | G | R | Y | V | S | Q | I | R | A | Q | L | V | K | V | V | F | Q | - | G | I | E | P | Q | I | N | - | D | W | V | A | I | T | K | V | K | K | G | G | A | N | K | V | V | D | S | G | Y | I | P | A | Y | L | L | P | K | V | Q | P | E | L | Q | W | A | W | T | N | L | D | E | Y | K | L | A | A | L | N | L | E | E | R | K | R | L | V | A | Q | F | L | A | E | S |
| - | - | M | M | S | I | A | Q | V | R | - | - | S | A | G | S | A | G | N | Y | Y | T | D | K | D | N | Y | Y | V | L | G | S | M | G | E | R | W | A | G | R | G | A | E | Q | L | G | L | Q | G | S | V | D | K | D | V | F | T | R | L | L | E | G | R | L | P | - | - | D | G | A | D | L | S | R | M | Q | D | G | S | N | R | H | R | P | G | Y | D | L | T | F | S | A | P | K | S | V | S | M | M | A | M | L | G | G | D | K | R | L | I | D | A | H | N | Q | A | V | D | F | A | V | R | Q | V | E | A | L | A | S | T | R | V | M | T | D | G | Q | S | E | T | V | L | T | G | N | L | V | M | A | L | F | N | H | D | T | S | R | D | Q | E | P | Q | L | H | T | H | A | V | V | A | N | V | T | Q | H | N | - | - | - | - | - | - | - | - | - | - | - | G | E | W | K | T | L | - | S | S | D | K | V | G | K | T | G | F | I | E | N | V | Y | A | N | Q | I | A | F | G | R | L | Y | R | E | K | L | K | E | Q | V | E | A | L | G | Y | E | T | E | V | V | G | K | H | G | M | W | E | M | P | G | V | P | V | E | A | F | S | V | D | P | E | I | K | M | A | E | W | M | Q | T | - | L | K | E | T | G | F | D | - | - | - | - | - | - | - | - | I | R | A | Y | R | D | A | A | D | Q | R | A | D | L | R | T | L | T | P | G |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | Q | G | A | M | L | A | V | F | K | D | T | Y | G | L | S | F | T | D | - | - | L | V | R | - | - | - | - | - | - | - | T | C | T | D | W | V | T | A | I | F | G | V | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | T | I | A | E | G | F | K | T | L | I | Q | P | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | L | Y | A | H | I | Q | C | L | D | C | K | - | - | - | - | - | W | G | V | L | I | L | A | L | L | R | Y | K | C | G | K | S | R | L | T | V | A | K | G | L | S | T | L | L | H | V | - | - | P | E | T | C | M | L | I | Q | P | P | - | - | K | L | - | - | - | - | - | R | S | S | V | A | A | L | Y | W | Y | R | T | G | I | S | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | I | S | E | V | M | G | - | - | - | - | - | D | T | P | E | W | I | Q | R | L | T | I | I | Q | - | - | - | - | - | - | - | - | - |
| - | - | M | L | D | I | T | T | I | T | R | Q | N | V | T | S | V | V | G | Y | Y | S | D | A | D | D | Y | Y | S | - | - | S | S | F | T | S | W | Q | G | T | G | A | E | A | L | G | L | S | G | D | V | E | S | A | R | F | K | E | L | L | V | G | E | I | D | - | - | T | F | T | H | M | Q | R | - | - | - | G | A | K | K | E | R | L | G | Y | D | L | T | F | S | A | P | K | G | V | S | M | Q | A | L | I | H | G | D | K | T | I | I | E | A | H | E | K | A | V | A | A | A | V | R | E | A | E | K | L | A | Q | A | R | T | G | - | - | - | - | S | V | T | Q | N | T | N | N | L | V | V | A | T | F | R | H | E | T | - | - | - | R | D | P | D | L | H | T | H | A | F | V | M | N | M | T | Q | R | E | D | - | - | - | - | - | - | - | - | - | - | G | Q | W | R | A | L | - | K | - | - | - | - | - | - | - | - | - | N | D | E | L | M | R | N | K | M | H | L | G | D | V | Y | K | Q | E | L | A | L | E | L | T | K | A | G | Y | E | L | R | Y | N | S | N | - | N | T | F | D | M | A | H | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | P | D | F | P | V | D | L | H | A | F | - | - | L | S | Q | A | V | - | - | F | - | - | - | - | - | - | - | - | - | S | N | T | V | A | S | F | A | V | Y | T | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | E | K | A | Q | I | L | Y | K | K | L | M | E | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | - | S | V | T | F | I | S | R | H | G | F | G | - | - | - | - | - | G | - | - | H | N | I | L | F | F | L | T | P | H | R | H | R | V | S | A | I | N | N | Y | C | Q | K | L | C | T | - | - | - | - | F | S | F | L | I | C | K | G | V | - | - | N | - | - | - | - | - | - | - | - | - | - | K | E | Y | L | F | Y | S | A | L | C | R | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | Y | A | V | V | E | E | S | I | Q | G | G | L | K | E | H | D | F | N | P | E | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | F | R | V | Y | S | K | Y | L | F | L | T | Y | P | Q | C | T | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | P | Q | Y | A | L | D | S | L | R | T | L | L | N | K | Y | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | P | L | Y | I | A | A | V | R | E | L | H | E | - | - | - | - | D | G | S | P | H | L | H | V | L | V | Q | N | L | R | A | S | I | - | - | T | N | P | N | A | L | N | - | - | L | R | M | D | T | S | P | F | S | I | F | H | P | N | I | Q | A | A | K | - | - | - | - | - | D | C | N | Q | V | R | D | Y | I | T | K | E | V | D | S | D | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | T | A | E | W | G | T | F | V | A | V | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1r9wa_ | A | P06789 | GO:0003677 |
