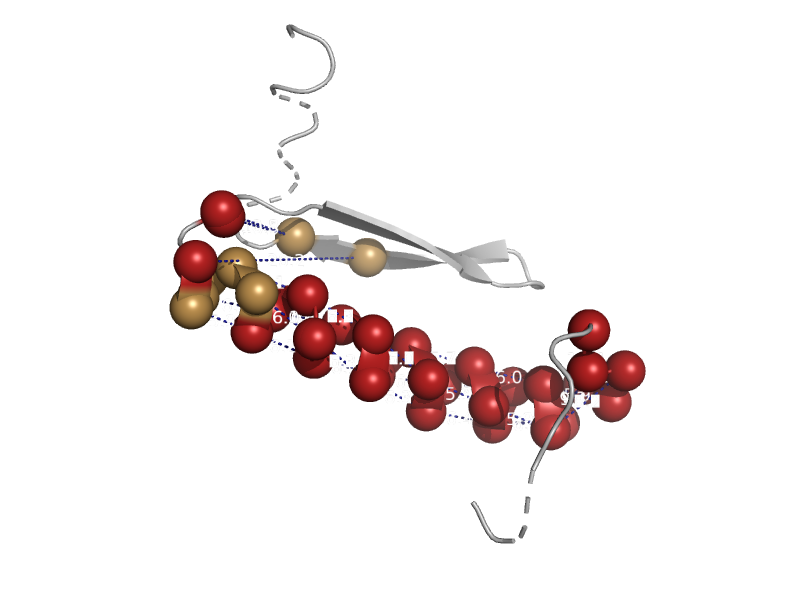
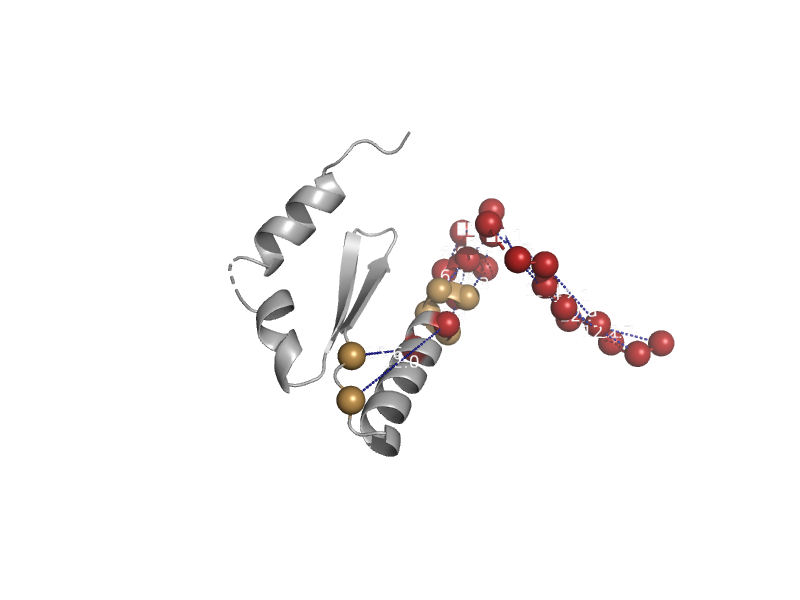
solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1egwa- ( 2 ) g---rkiqitrimdernrqvtftkrkfglmkkayelsvlcdceialiifn
d1mnma- ( 15 ) qkerrkiei-fienktrrhvtfskrkhgimkkafelsvltgtqvlllvvs
aaaaaaaaaaaaaaaaaaaaaaaaa bbbbb
60 70 80
d1egwa- ( 50 ) ssnklfqyastmvlky-tey
d1mnma- ( 65 ) etglvytfStpkfepivtqegrnliqaclnapdd
bbbbb
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | - | - | - | R | K | I | Q | I | T | R | I | M | D | E | R | N | R | Q | V | T | F | T | K | R | K | F | G | L | M | K | K | A | Y | E | L | S | V | L | C | D | C | E | I | A | L | I | I | F | N | S | S | N | K | L | F | Q | Y | A | S | T | M | V | L | K | Y | - | T | E | Y | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Q | K | E | R | R | K | I | E | I | - | F | I | E | N | K | T | R | R | H | V | T | F | S | K | R | K | H | G | I | M | K | K | A | F | E | L | S | V | L | T | G | T | Q | V | L | L | L | V | V | S | E | T | G | L | V | Y | T | F | S | T | P | K | F | E | P | I | V | T | Q | E | G | R | N | L | I | Q | A | C | L | N | A | P | D | D |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1egwa_ | A | Q02078 | GO:0000977 |
| d1egwa_ | A | Q02078 | GO:0003677 |
| d1egwa_ | A | Q02078 | GO:0045944 |
| d1egwa_ | A | Q02078 | GO:0046983 |
| d1mnma_ | A | P11746 | GO:0000981 |
| d1mnma_ | A | P11746 | GO:0000987 |
| d1mnma_ | A | P11746 | GO:0003677 |
| d1mnma_ | A | P11746 | GO:0045944 |
| d1mnma_ | A | P11746 | GO:0046983 |
| d1mnma_ | A | P0CY08 | GO:0000981 |
| d1mnma_ | A | P0CY08 | GO:0000987 |
| d1mnma_ | A | P0CY08 | GO:0003677 |
| d1mnma_ | A | P0CY08 | GO:0045944 |
| d1mnma_ | A | P0CY08 | GO:0046983 |