solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1w5fa2 ( 216 ) yirltsrfariesVmk--dAgaailGigvGkg-------ehRAreAAkkA
d2btoa2 ( 253 ) e---islrelltnLvpqpsLhflMcAfApltppd-----elgieeMIksL
d2r7512 ( 205 ) vdfadvrtTle--eGglsIiGmgeGrg-------dekAdiAVekA
d2vapa2 ( 232 ) invdfadvkaVmn--ngglamiGiGesds-------ekrAkeAVsmA
d3cb2a2 ( 247 ) gymnndligliasLiptprLhflMtGytplt-------vrttVldVMrrL
d3ryca2 ( 246 ) galNvdltefqtnLvpyprIhfpLaTyAPvisaekayheqlsVaeITnaC
d3rycb2 ( 246 ) gqlnadlrklavnMvpfprLhffMpGfAPltsrg----yaLtVpeLTqqM
d6rvqa2 ( 209 ) ldfadvktims--nqgsalmgiGvssg-------enrAveAAkkA
aaaaaaa bbbbb aaaaaaa
60 70 80 90 100
d1w5fa2 ( 257 ) Mesklie--hpVenAssIvFnItAPsnIrmeEvheAamIIrqnSs----e
d2btoa2 ( 300 ) FdngsVfaaCsPmeGrFLSTAVlyrGi---pladaaLaamrek--LplTy
d2r7512 ( 241 ) Vtsplle-gntIegArrLlVtIwTSedipydivdeVmerIhskVh----p
d2vapa2 ( 270 ) lnSplLd--VdidgAtgAlIhVmGPedLtleeAreVvatVssrLd----p
d3cb2a2 ( 298 ) Lq-pnvmvstg-tnhCyIAIlNiIqg-vdptqvh-slqIr----lAnfIp
d3ryca2 ( 296 ) fepanQmvkCdprhgkyMaCCLlYrGdVvpkdVnaAiatiktkrsiqFVd
d3rycb2 ( 296 ) fdskNMmaaCdPrhGrYLtVATiFrGrmsmkeVdeqMlniqnknssyfVe
d6rvqa2 ( 245 ) issplle--tsivgAqgvlMnitGGeslslfeaqeAadivqdaAd----e
333 bbbbbbbb aaaaaaaaaaaa
110 120 130 140 150
d1w5fa2 ( 301 ) dAdvkfglifdd----------eVpddeIrViFiATrFpde----dkil-
d2btoa2 ( 349 ) wiptaFkigyveqpgi--------shrKSMvLlANNTeiar----vldri
d2r7512 ( 286 ) eAeIifgavlep----------qe-qdFIrVaIvATdFpeekfqv--ge-
d2vapa2 ( 314 ) nAtiiwgatide----------nl-entVrVlLvITgVqsrIeftdt---
d3cb2a2 ( 351 ) wgpasI-Valsrsp------------yvsGlMmANhTsISs----lfe--
d3ryca2 ( 346 ) wcptgFkvginyqpptvvpggdlakVqrAVcMlSNTtAiae----awarl
d3rycb2 ( 346 ) wipnnVktavcdippr--------glkmSStFiGNSTAIQe----lfkri
d6rvqa2 ( 289 ) dvnmifgtvinp----------el-qdeivvtviAtgf
bbbbb bbbbbbbbb
160 170 180 190 200
d1w5fa2 ( 336 ) --------------------f
d2btoa2 ( 387 ) Chnfdklwqrkafanwylnegms--eeqinvlrasaqelVqsYqvAee
d2r7512 ( 322 ) --------------------kevkf-------------------------
d2vapa2 ( 350 ) --------------------glkrk
d3cb2a2 ( 391 ) tcqydl--rreafleqfrkedmfdnfdemdtsr-ivq-lIdeYhaAtrpd
d3ryca2 ( 392 ) DhkfdlmyakrafVhwyvgegme--egefsearedmaalekdYeevgvd
d3rycb2 ( 392 ) seqftamfrrkaflhwytgegmd--emefteaesnmndlvseYqqyqdat
d6rvqa2
d1w5fa2
d2btoa2
d2r7512 ( 327 ) kvik
d2vapa2
d3cb2a2 ( 443 ) yisw
d3ryca2
d3rycb2 ( 440 ) -ade
d6rvqa2
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Cell-division protein FtsZ | Tubulin, C-terminal domain | Thermotoga maritima [TaxId: 2336] | 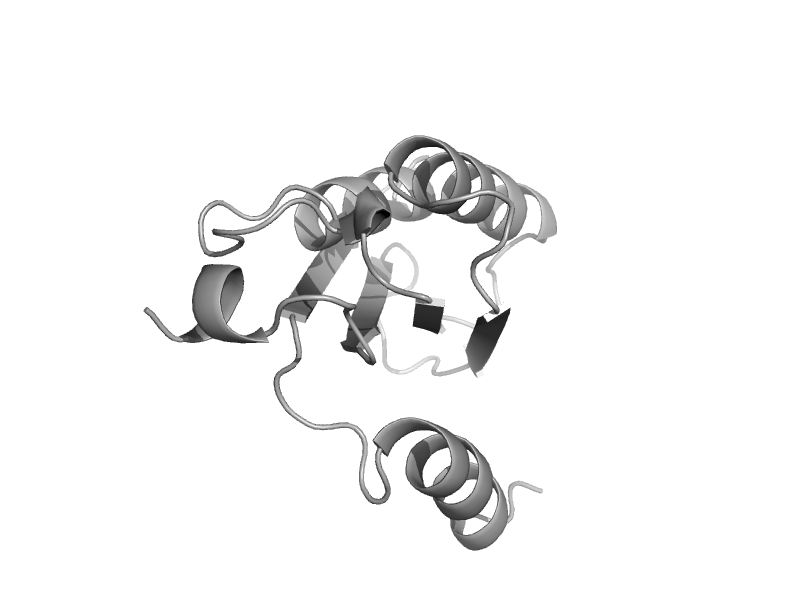 | view | |
| Tubulin alpha-subunit | Tubulin, C-terminal domain | Prosthecobacter dejongeii [TaxId: 48465] | 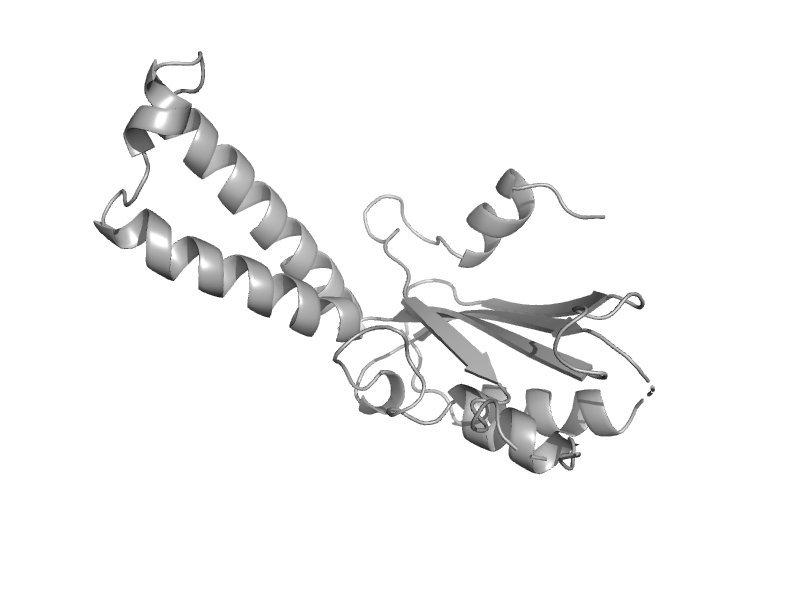 | view | |
| Cell-division protein FtsZ | Tubulin, C-terminal domain | Aquifex aeolicus [TaxId: 63363] | 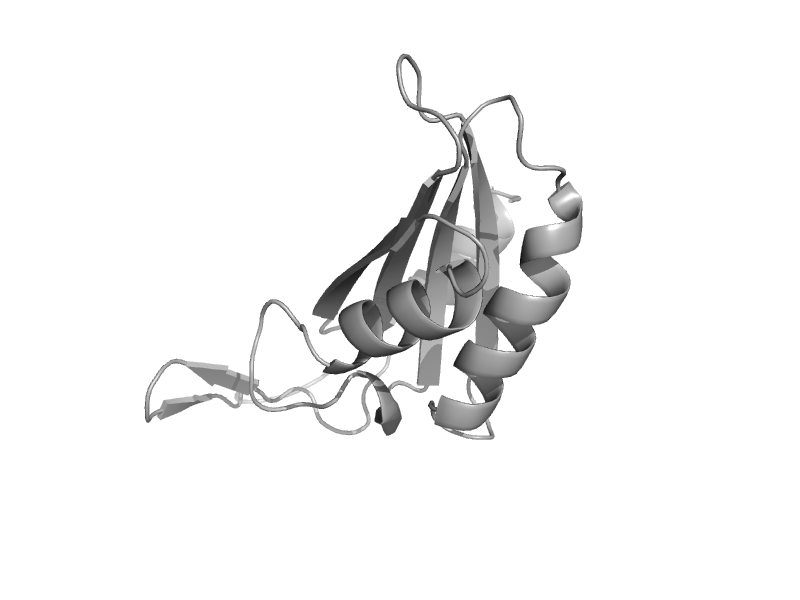 | view | |
| Cell-division protein FtsZ | Tubulin, C-terminal domain | Methanococcus jannaschii [TaxId: 2190] | 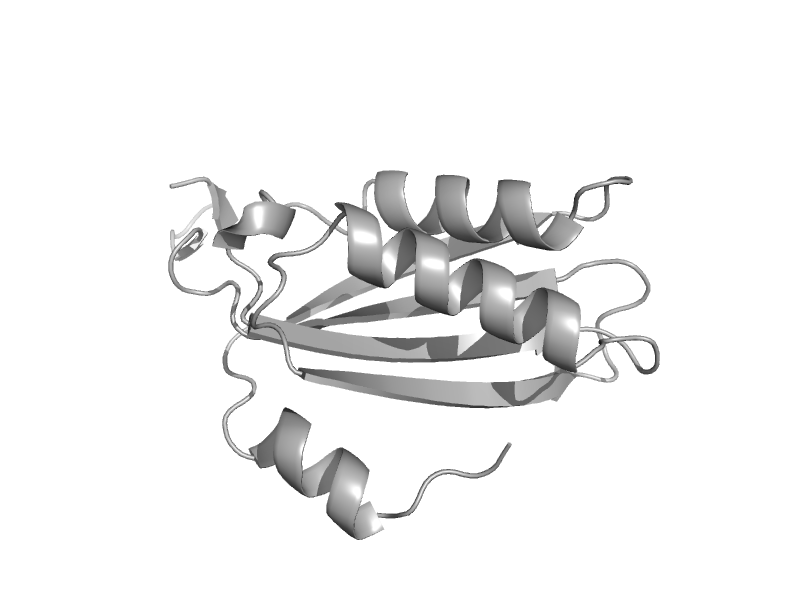 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 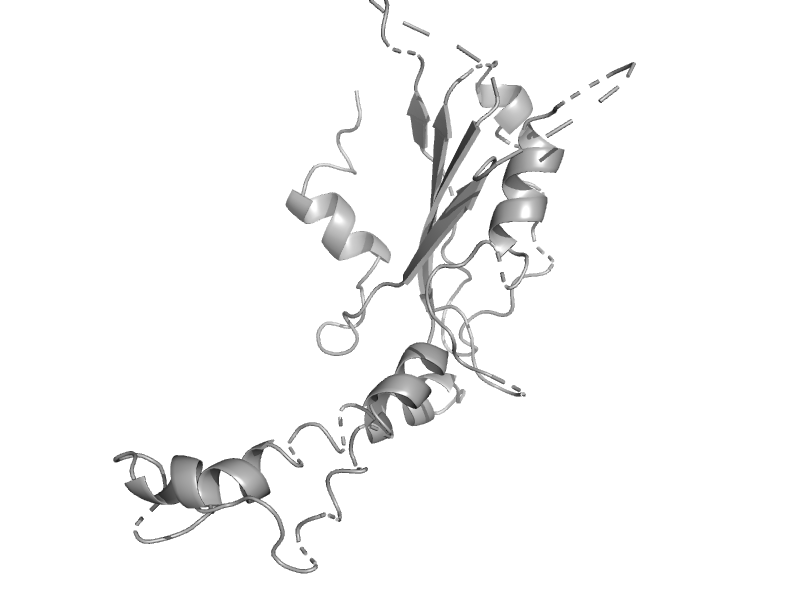 | view | |
| automated matches | Tubulin, C-terminal domain | Sheep (Ovis aries) [TaxId: 9940] | 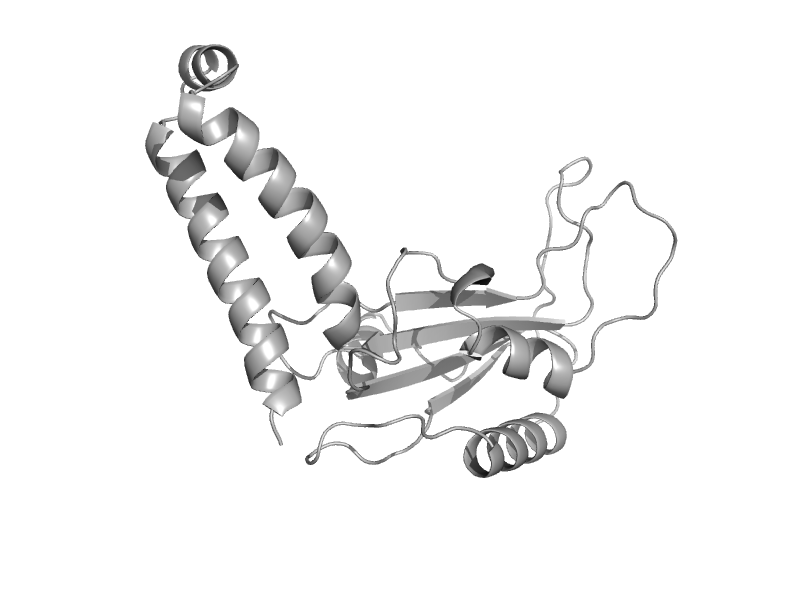 | view | |
| automated matches | Tubulin, C-terminal domain | Sheep (Ovis aries) [TaxId: 9940] | 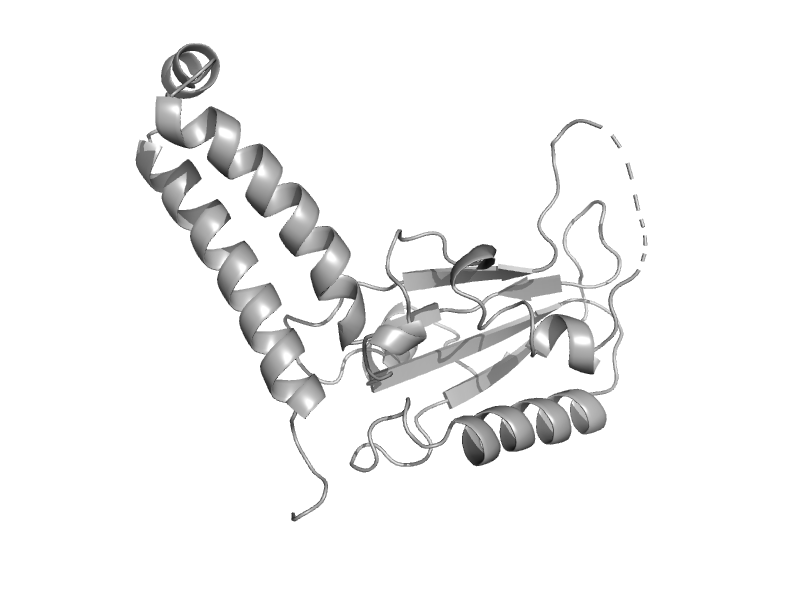 | view | |
| automated matches | automated matches | Staphylococcus aureus [TaxId: 1280] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Y | I | R | L | T | S | R | F | A | R | I | E | S | V | M | K | - | - | D | A | G | A | A | I | L | G | I | G | V | G | K | G | - | - | - | - | - | - | - | E | H | R | A | R | E | A | A | K | K | A | M | E | S | K | L | I | E | - | - | H | P | V | E | N | A | S | S | I | V | F | N | I | T | A | P | S | N | I | R | M | E | E | V | H | E | A | A | M | I | I | R | Q | N | S | S | - | - | - | - | E | D | A | D | V | K | F | G | L | I | F | D | D | - | - | - | - | - | - | - | - | - | - | E | V | P | D | D | E | I | R | V | I | F | I | A | T | R | F | P | D | E | - | - | - | - | D | K | I | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| E | - | - | - | I | S | L | R | E | L | L | T | N | L | V | P | Q | P | S | L | H | F | L | M | C | A | F | A | P | L | T | P | P | D | - | - | - | - | - | E | L | G | I | E | E | M | I | K | S | L | F | D | N | G | S | V | F | A | A | C | S | P | M | E | G | R | F | L | S | T | A | V | L | Y | R | G | I | - | - | - | P | L | A | D | A | A | L | A | A | M | R | E | K | - | - | L | P | L | T | Y | W | I | P | T | A | F | K | I | G | Y | V | E | Q | P | G | I | - | - | - | - | - | - | - | - | S | H | R | K | S | M | V | L | L | A | N | N | T | E | I | A | R | - | - | - | - | V | L | D | R | I | C | H | N | F | D | K | L | W | Q | R | K | A | F | A | N | W | Y | L | N | E | G | M | S | - | - | E | E | Q | I | N | V | L | R | A | S | A | Q | E | L | V | Q | S | Y | Q | V | A | E | E | - | - | - | - | - | - |
| - | - | - | - | - | V | D | F | A | D | V | R | T | T | L | E | - | - | E | G | G | L | S | I | I | G | M | G | E | G | R | G | - | - | - | - | - | - | - | D | E | K | A | D | I | A | V | E | K | A | V | T | S | P | L | L | E | - | G | N | T | I | E | G | A | R | R | L | L | V | T | I | W | T | S | E | D | I | P | Y | D | I | V | D | E | V | M | E | R | I | H | S | K | V | H | - | - | - | - | P | E | A | E | I | I | F | G | A | V | L | E | P | - | - | - | - | - | - | - | - | - | - | Q | E | - | Q | D | F | I | R | V | A | I | V | A | T | D | F | P | E | E | K | F | Q | V | - | - | G | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | E | V | K | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | V | I | K |
| - | - | - | I | N | V | D | F | A | D | V | K | A | V | M | N | - | - | N | G | G | L | A | M | I | G | I | G | E | S | D | S | - | - | - | - | - | - | - | E | K | R | A | K | E | A | V | S | M | A | L | N | S | P | L | L | D | - | - | V | D | I | D | G | A | T | G | A | L | I | H | V | M | G | P | E | D | L | T | L | E | E | A | R | E | V | V | A | T | V | S | S | R | L | D | - | - | - | - | P | N | A | T | I | I | W | G | A | T | I | D | E | - | - | - | - | - | - | - | - | - | - | N | L | - | E | N | T | V | R | V | L | L | V | I | T | G | V | Q | S | R | I | E | F | T | D | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | L | K | R | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| G | Y | M | N | N | D | L | I | G | L | I | A | S | L | I | P | T | P | R | L | H | F | L | M | T | G | Y | T | P | L | T | - | - | - | - | - | - | - | V | R | T | T | V | L | D | V | M | R | R | L | L | Q | - | P | N | V | M | V | S | T | G | - | T | N | H | C | Y | I | A | I | L | N | I | I | Q | G | - | V | D | P | T | Q | V | H | - | S | L | Q | I | R | - | - | - | - | L | A | N | F | I | P | W | G | P | A | S | I | - | V | A | L | S | R | S | P | - | - | - | - | - | - | - | - | - | - | - | - | Y | V | S | G | L | M | M | A | N | H | T | S | I | S | S | - | - | - | - | L | F | E | - | - | T | C | Q | Y | D | L | - | - | R | R | E | A | F | L | E | Q | F | R | K | E | D | M | F | D | N | F | D | E | M | D | T | S | R | - | I | V | Q | - | L | I | D | E | Y | H | A | A | T | R | P | D | Y | I | S | W |
| G | A | L | N | V | D | L | T | E | F | Q | T | N | L | V | P | Y | P | R | I | H | F | P | L | A | T | Y | A | P | V | I | S | A | E | K | A | Y | H | E | Q | L | S | V | A | E | I | T | N | A | C | F | E | P | A | N | Q | M | V | K | C | D | P | R | H | G | K | Y | M | A | C | C | L | L | Y | R | G | D | V | V | P | K | D | V | N | A | A | I | A | T | I | K | T | K | R | S | I | Q | F | V | D | W | C | P | T | G | F | K | V | G | I | N | Y | Q | P | P | T | V | V | P | G | G | D | L | A | K | V | Q | R | A | V | C | M | L | S | N | T | T | A | I | A | E | - | - | - | - | A | W | A | R | L | D | H | K | F | D | L | M | Y | A | K | R | A | F | V | H | W | Y | V | G | E | G | M | E | - | - | E | G | E | F | S | E | A | R | E | D | M | A | A | L | E | K | D | Y | E | E | V | G | V | D | - | - | - | - | - |
| G | Q | L | N | A | D | L | R | K | L | A | V | N | M | V | P | F | P | R | L | H | F | F | M | P | G | F | A | P | L | T | S | R | G | - | - | - | - | Y | A | L | T | V | P | E | L | T | Q | Q | M | F | D | S | K | N | M | M | A | A | C | D | P | R | H | G | R | Y | L | T | V | A | T | I | F | R | G | R | M | S | M | K | E | V | D | E | Q | M | L | N | I | Q | N | K | N | S | S | Y | F | V | E | W | I | P | N | N | V | K | T | A | V | C | D | I | P | P | R | - | - | - | - | - | - | - | - | G | L | K | M | S | S | T | F | I | G | N | S | T | A | I | Q | E | - | - | - | - | L | F | K | R | I | S | E | Q | F | T | A | M | F | R | R | K | A | F | L | H | W | Y | T | G | E | G | M | D | - | - | E | M | E | F | T | E | A | E | S | N | M | N | D | L | V | S | E | Y | Q | Q | Y | Q | D | A | T | - | A | D | E |
| - | - | - | - | - | L | D | F | A | D | V | K | T | I | M | S | - | - | N | Q | G | S | A | L | M | G | I | G | V | S | S | G | - | - | - | - | - | - | - | E | N | R | A | V | E | A | A | K | K | A | I | S | S | P | L | L | E | - | - | T | S | I | V | G | A | Q | G | V | L | M | N | I | T | G | G | E | S | L | S | L | F | E | A | Q | E | A | A | D | I | V | Q | D | A | A | D | - | - | - | - | E | D | V | N | M | I | F | G | T | V | I | N | P | - | - | - | - | - | - | - | - | - | - | E | L | - | Q | D | E | I | V | V | T | V | I | A | T | G | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1w5fa2 | A | O08398 | GO:0003924 |
| d1w5fa2 | A | O08398 | GO:0005525 |
| d2btoa2 | A | Q8GCC5 | GO:0000226 |
| d2btoa2 | A | Q8GCC5 | GO:0005200 |
| d2btoa2 | A | Q8GCC5 | GO:0005515 |
| d2btoa2 | A | Q8GCC5 | GO:0005525 |
| d2btoa2 | A | Q8GCC5 | GO:0005737 |
| d2btoa2 | A | Q8GCC5 | GO:0005874 |
| d2btoa2 | A | Q8GCC5 | GO:0007017 |
| d2btoa2 | A | Q8GCC5 | GO:0016787 |
| d2btoa2 | A | P0AA25 | GO:0000226 |
| d2btoa2 | A | P0AA25 | GO:0005200 |
| d2btoa2 | A | P0AA25 | GO:0005515 |
| d2btoa2 | A | P0AA25 | GO:0005525 |
| d2btoa2 | A | P0AA25 | GO:0005737 |
| d2btoa2 | A | P0AA25 | GO:0005874 |
| d2btoa2 | A | P0AA25 | GO:0007017 |
| d2btoa2 | A | P0AA25 | GO:0016787 |
| d2vapa2 | A | Q57816 | GO:0000917 |
| d2vapa2 | A | Q57816 | GO:0003924 |
| d2vapa2 | A | Q57816 | GO:0005525 |
| d2vapa2 | A | Q57816 | GO:0005737 |
| d2vapa2 | A | Q57816 | GO:0032153 |
| d2vapa2 | A | Q57816 | GO:0043093 |
| d2vapa2 | A | Q57816 | GO:0051258 |
| d2vapa2 | A | Q57816 | GO:0051301 |
| d2vapa2 | A | Q57816 | GO:0090529 |
