solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1d5ya3 ( 122 ) eftmpehkfvtledtpLiGvtqsyscsleq
d1jyha- ( 1 ) mnyeikqeekrtVAGfhlvg--pwe-
d1r8ea2 ( 121 ) lgeVfvldeeeirIIqteAegigPenv
d2gova1 ( 7 ) nslfgsvetwpwqvl--stgGkedvsyeeraCEgGkFAtvevtdkpvdeA
d3r8ja- ( 19 ) avetPgwkapedagpqpg--sYeiRhygpakWVSTsvesmdwdsA
bbbbb bbbbbbbbb
60 70 80 90 100
d1d5ya3 ( 152 ) isdfRhemryqfwhdFlgnAptiPpvLyGLnetrpsqdkddeqevfYTTA
d1jyha- ( 24 ) --qtvkkGfeqLmmwVdsk-nivpkewVAVyydnpdetpaek--LrCDTV
d1r8ea2 ( 148 ) lnasYskLkkfIesaDg----ftNnsyGATFsfqpytside--MtYrhIF
d2gova1 ( 55 ) lreAmpkimkYvggtndkgvgmgMtvpvSfavfPne---dgslqkklkvw
d3r8ja- ( 62 ) iqtGftkLnsYiqgkNekemkIkmTAPVTSyvepgs--gpfs-estItIS
aaaaaaaaaaa bbbb bbbbb
110 120 130 140 150
d1d5ya3 ( 202 ) La-qdqA-----dgyVltGhpvmLqggeYVmFtyegl--gtgvqeFiltV
d1jyha- ( 69 ) VTVpgyf---tlpenSegVilteItggqYAVAvarVv--gddfakPwyqF
d1r8ea2 ( 192 ) Tpvltn---kqissitpdMeittIpkgrYAcIaYnfs--pehYflNLqkL
d2gova1 ( 102 ) FriPnqfqGspPapsdesvkieeregitvYStqfggyAkeadyvaHAtql
d3r8ja- ( 109 ) LyIPseqqfdPPrPlesdVfiedraemtVFVrsFdgfSsaqkNqeqlltL
bb bbbbb bbbbbbbb aaaaaaaa
160 170 180 190
d1d5ya3 ( 244 ) ygtCmpmln-ltrrkgQdIEryypaeddrpinlrCeLLIPIrr
d1jyha- ( 114 ) fn-sLlqdsayemlpkpcFEVylnnGaedg-ywdIeMYVAVqpk
d1r8ea2 ( 237 ) ik-yIadrq-ltvvsdVyEliipihyspkqeeyrVeMKIrIa
d2gova1 ( 152 ) rt-tlegt-patyqgdvYyCAgydppmkpygrrnEvwlvka
d3r8ja- ( 159 ) as-iLred-gkvFdekvyYTAgYnspvkllnrnNEVWliqk--n
aa aa bbbbb bbbbbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Rob transcription factor, C-terminal domain | Rob transcription factor, C-terminal domain | Escherichia coli [TaxId: 562] | 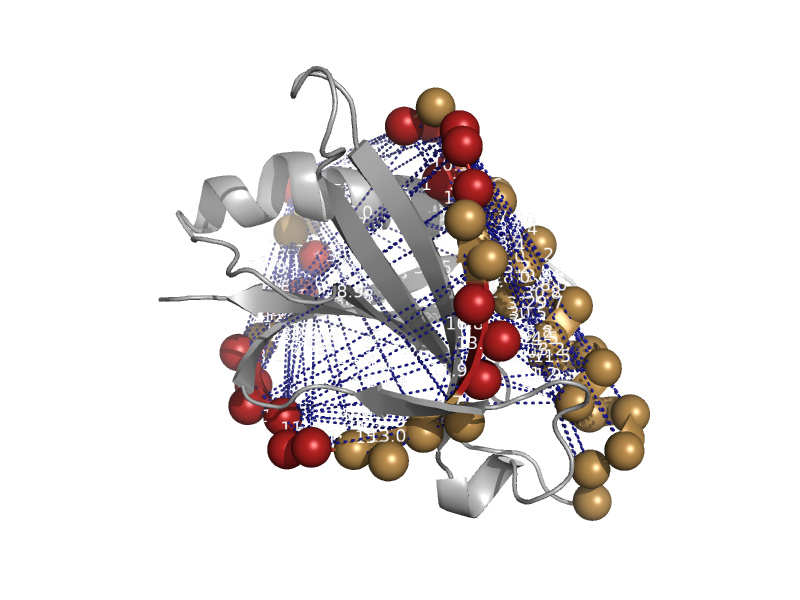 | view | |
| Gyrase inhibitory protein GyrI (SbmC, YeeB) | Gyrase inhibitory protein GyrI (SbmC, YeeB) | Escherichia coli [TaxId: 562] |  | view | |
| Multidrug-binding domain of transcription activator BmrR | Multidrug-binding domain of transcription activator BmrR | Bacillus subtilis [TaxId: 1423] | 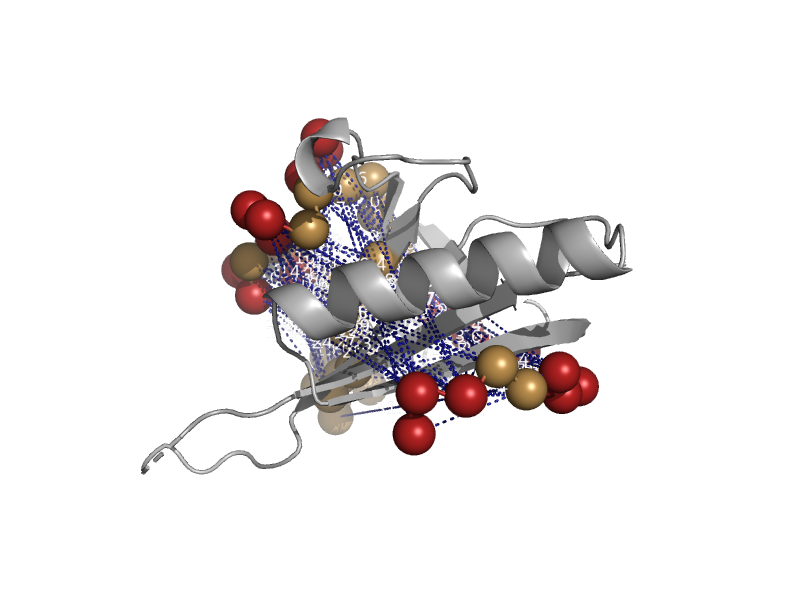 | view | |
| Heme-binding protein 1 | SOUL heme-binding protein | Mouse (Mus musculus) [TaxId: 10090] | 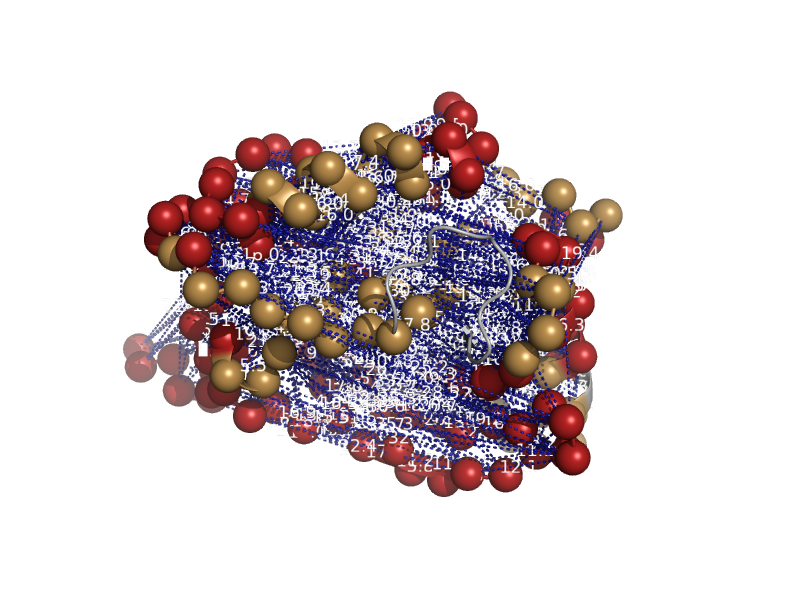 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 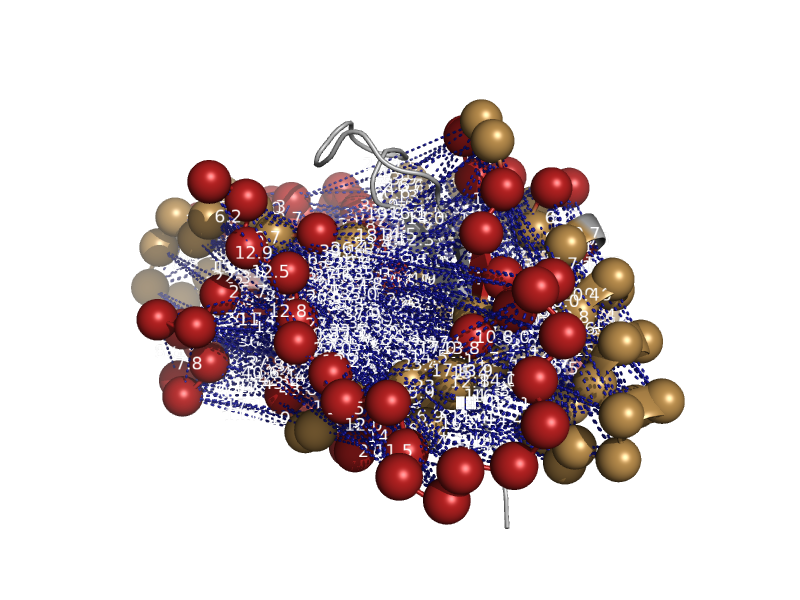 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | F | T | M | P | E | H | K | F | V | T | L | E | D | T | P | L | I | G | V | T | Q | S | Y | S | C | S | L | E | Q | I | S | D | F | R | H | E | M | R | Y | Q | F | W | H | D | F | L | G | N | A | P | T | I | P | P | V | L | Y | G | L | N | E | T | R | P | S | Q | D | K | D | D | E | Q | E | V | F | Y | T | T | A | L | A | - | Q | D | Q | A | - | - | - | - | - | D | G | Y | V | L | T | G | H | P | V | M | L | Q | G | G | E | Y | V | M | F | T | Y | E | G | L | - | - | G | T | G | V | Q | E | F | I | L | T | V | Y | G | T | C | M | P | M | L | N | - | L | T | R | R | K | G | Q | D | I | E | R | Y | Y | P | A | E | D | D | R | P | I | N | L | R | C | E | L | L | I | P | I | R | R | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | N | Y | E | I | K | Q | E | E | K | R | T | V | A | G | F | H | L | V | G | - | - | P | W | E | - | - | - | Q | T | V | K | K | G | F | E | Q | L | M | M | W | V | D | S | K | - | N | I | V | P | K | E | W | V | A | V | Y | Y | D | N | P | D | E | T | P | A | E | K | - | - | L | R | C | D | T | V | V | T | V | P | G | Y | F | - | - | - | T | L | P | E | N | S | E | G | V | I | L | T | E | I | T | G | G | Q | Y | A | V | A | V | A | R | V | V | - | - | G | D | D | F | A | K | P | W | Y | Q | F | F | N | - | S | L | L | Q | D | S | A | Y | E | M | L | P | K | P | C | F | E | V | Y | L | N | N | G | A | E | D | G | - | Y | W | D | I | E | M | Y | V | A | V | Q | P | K |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | G | E | V | F | V | L | D | E | E | E | I | R | I | I | Q | T | E | A | E | G | I | G | P | E | N | V | L | N | A | S | Y | S | K | L | K | K | F | I | E | S | A | D | G | - | - | - | - | F | T | N | N | S | Y | G | A | T | F | S | F | Q | P | Y | T | S | I | D | E | - | - | M | T | Y | R | H | I | F | T | P | V | L | T | N | - | - | - | K | Q | I | S | S | I | T | P | D | M | E | I | T | T | I | P | K | G | R | Y | A | C | I | A | Y | N | F | S | - | - | P | E | H | Y | F | L | N | L | Q | K | L | I | K | - | Y | I | A | D | R | Q | - | L | T | V | V | S | D | V | Y | E | L | I | I | P | I | H | Y | S | P | K | Q | E | E | Y | R | V | E | M | K | I | R | I | A | - | - |
| N | S | L | F | G | S | V | E | T | W | P | W | Q | V | L | - | - | S | T | G | G | K | E | D | V | S | Y | E | E | R | A | C | E | G | G | K | F | A | T | V | E | V | T | D | K | P | V | D | E | A | L | R | E | A | M | P | K | I | M | K | Y | V | G | G | T | N | D | K | G | V | G | M | G | M | T | V | P | V | S | F | A | V | F | P | N | E | - | - | - | D | G | S | L | Q | K | K | L | K | V | W | F | R | I | P | N | Q | F | Q | G | S | P | P | A | P | S | D | E | S | V | K | I | E | E | R | E | G | I | T | V | Y | S | T | Q | F | G | G | Y | A | K | E | A | D | Y | V | A | H | A | T | Q | L | R | T | - | T | L | E | G | T | - | P | A | T | Y | Q | G | D | V | Y | Y | C | A | G | Y | D | P | P | M | K | P | Y | G | R | R | N | E | V | W | L | V | K | A | - | - | - |
| - | - | - | - | - | A | V | E | T | P | G | W | K | A | P | E | D | A | G | P | Q | P | G | - | - | S | Y | E | I | R | H | Y | G | P | A | K | W | V | S | T | S | V | E | S | M | D | W | D | S | A | I | Q | T | G | F | T | K | L | N | S | Y | I | Q | G | K | N | E | K | E | M | K | I | K | M | T | A | P | V | T | S | Y | V | E | P | G | S | - | - | G | P | F | S | - | E | S | T | I | T | I | S | L | Y | I | P | S | E | Q | Q | F | D | P | P | R | P | L | E | S | D | V | F | I | E | D | R | A | E | M | T | V | F | V | R | S | F | D | G | F | S | S | A | Q | K | N | Q | E | Q | L | L | T | L | A | S | - | I | L | R | E | D | - | G | K | V | F | D | E | K | V | Y | Y | T | A | G | Y | N | S | P | V | K | L | L | N | R | N | N | E | V | W | L | I | Q | K | - | - | N |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1d5ya3 | A | P0ACI0 | GO:0003677 |
| d1d5ya3 | A | P0ACI0 | GO:0003700 |
| d1d5ya3 | A | P0ACI0 | GO:0005829 |
| d1d5ya3 | A | P0ACI0 | GO:0006355 |
| d1d5ya3 | A | P0ACI0 | GO:0043565 |
| d1d5ya3 | A | P0ACI0 | GO:0003677 |
| d1d5ya3 | A | P0ACI0 | GO:0003700 |
| d1d5ya3 | A | P0ACI0 | GO:0005829 |
| d1d5ya3 | A | P0ACI0 | GO:0006355 |
| d1d5ya3 | A | P0ACI0 | GO:0043565 |
| d1jyha_ | A | P33012 | GO:0005737 |
| d1jyha_ | A | P33012 | GO:0006974 |
| d1jyha_ | A | P33012 | GO:0008657 |
| d1jyha_ | A | P33012 | GO:2000104 |
| d1jyha_ | A | P33012 | GO:2000372 |
| d1r8ea2 | A | P39075 | GO:0003677 |
| d1r8ea2 | A | P39075 | GO:0003700 |
| d1r8ea2 | A | P39075 | GO:0006355 |
| d1r8ea2 | A | P39075 | GO:0003677 |
| d1r8ea2 | A | P39075 | GO:0003700 |
| d1r8ea2 | A | P39075 | GO:0006355 |
| d1r8ea2 | A | P39075 | GO:0003677 |
| d1r8ea2 | A | P39075 | GO:0003700 |
| d1r8ea2 | A | P39075 | GO:0006355 |
| d2gova1 | A | Q9R257 | GO:0005737 |
| d2gova1 | A | Q9R257 | GO:0005739 |
| d2gova1 | A | Q9R257 | GO:0005829 |
| d2gova1 | A | Q9R257 | GO:0020037 |
| d2gova1 | A | Q9R257 | GO:0042168 |
