solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1nh8a2 ( 211 ) qylmLdYdCp-rs------alkkAtaiTpGle---sPtiapl-
d1nzaa- ( 1 ) meevVlItVpsee------vArtIakaLveerlAAcVnivpgl
d1o2ca1 ( 2 ) klLkIyLge-dkhsg-pLfeyLv-rayelg--kgvtvyrgi
d1ukua- ( 1 ) MIiVyTtFpdwe------sAekVvktLlkerLIAcAnlre-h
d2cz4a1 ( 2 ) dlvpLklVTIvAe-sl------LekrLveeVkrlg-Akgytitp-a
d2dcla- ( 2 ) vevehwnTlrLrIyIgendkwegrpLykvIvekLremg-Iagatvyrgi
d2hfva1 ( 23 ) lrellrtndav------llsavgalldgad--ighlvldq-
d2nuha- ( 3 ) sdVylIfSTCpdlp------sAeiIsrvLvqerlAAcVtqlpga
d2vd3a2 ( 213 ) gkrlVmLnId-rk------nldrVralMpgmt---gptvsev-
d3opka- ( 12 ) pAVvVlCtApdea------tAqdLaakVlaekLAAcAtllpga
d4affa1 ( 1 ) mkkIeAiIr-pf------kldeVkiaLvnag-Ivgmtvse-v
d4e98a- ( 6 ) tetkieSnIilIyIsApnqd------eAtsIaktLvdeeLCAcVsiipsv
d4iyqa1 ( 4 ) nISlLyTtTpty-------dAyrIsniLlenklIAcAnifs-i
d6mm2a1 ( 2 ) sqqvwklviitE-ei------llkkvskiikeAg-Asgytvla-a
d6t76a- ( 1 ) MkiAltnlp-pe------hGeriArllveehivAcvnlyp-v
bbbbbbb aaaaaaaaaa bbbb
60 70 80 90 100
d1nh8a2 ( 243 ) --------adp---------dwVaIrAlV-prrdvngimdeLaai-----
d1nzaa- ( 38 ) tsiyrwq-------gevvedqellLlVkT-tthAfpkLkerVkalHpyt-
d1o2ca1 ( 43 ) gfgh--------------pdlpIvLeivD-eeerInlFl----eidi---
d1ukua- ( 36 ) rafywwe-------gkieedkeVGAiLkT-redLweeLkerIkelHpyd-
d2cz4a1 ( 39 ) rge---gsrgirs--vdwegqnirLeTiV-seevAlrIlqrLqeeYfph-
d2dcla- ( 50 ) ygfg--------------tdlpIiVeVvd-rghnIekVvnvIkpmIk---
d2hfva1 ( 55 ) -------nmsileg----slgviprrvlv-heddlagArrlltdaglAhE
d2nuha- ( 41 ) vstyrwq-------gkiettqEiqLlIkT-NavhvnaAitrLcalHpyr-
d2vd3a2 ( 245 ) --------lsd--------ngVVaVhavV-dekevfnLinrLkav-----
d3opka- ( 50 ) tslyy-e-------gkleqeyEVqMiLkT-tvshqqaLidcLkshHpyq-
d4affa1 ( 34 ) rgfg----rqkgseytveflqklkLeIvV-edaqvdtVidkIvaaartge
d4e98a- ( 50 ) rsiykfk-------gqvhdeneVmLlVkT-TsqlfttLkekVteiHsye-
d4iyqa1 ( 41 ) tsvy-----------dihnntecaIiLkT-tndlvqhAtnkI-aiHpyd-
d6mm2a1 ( 38 ) agegsrnvrstgepsvshaysnikFevltasrelAdqiqdkvvakyfdd-
d6t76a- ( 35 ) hsiyswk-------gevcseaEvtlmMkv-StqGierlkqricelHpye-
bbbbbbbb aaaaaaaaaa
110 120 130
d1nh8a2 ( 270 ) -gAkailasdirfcrf
d1nzaa- ( 79 ) --vPeIvalpia----egnreyldwLrenTg
d1o2ca1 ( 87 ) --fglvftadvnvvk
d1ukua- ( 77 ) --vpaIiridvd----dvnedylkwLieeTkk
d2cz4a1 ( 82 ) -yaViAyvenv-wvv------rGekyv
d2dcla- ( 96 ) --dgmitveptivlwvgtq------------ee
d2hfva1 ( 93 ) lrsdd
d2nuha- ( 82 ) --lPeaiavqvs----vglpeyltwInteId
d2vd3a2 ( 273 ) -gArdilvvpieriip
d3opka- ( 91 ) --tPeLlvlpvt----hgdtdylswLnasLr
d4affa1 ( 86 ) ngdgkifvspvdqtirirtgeknadAi
d4e98a- ( 91 ) --lpeIiatkvv----ygnenyInwVnqtVr
d4iyqa1 ( 82 ) --tpaiitidpt----nan-dfiqwVndcTal
d6mm2a1 ( 87 ) -ysCityistv-eal
d6t76a- ( 76 ) --lpeFvvievdn--naslreyidfvkgethly
bbbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| ATP phosphoribosyltransferase (ATP-PRTase, HisG), regulatory C-terminal domain | ATP phosphoribosyltransferase (ATP-PRTase, HisG), regulatory C-terminal domain | Mycobacterium tuberculosis [TaxId: 1773] | 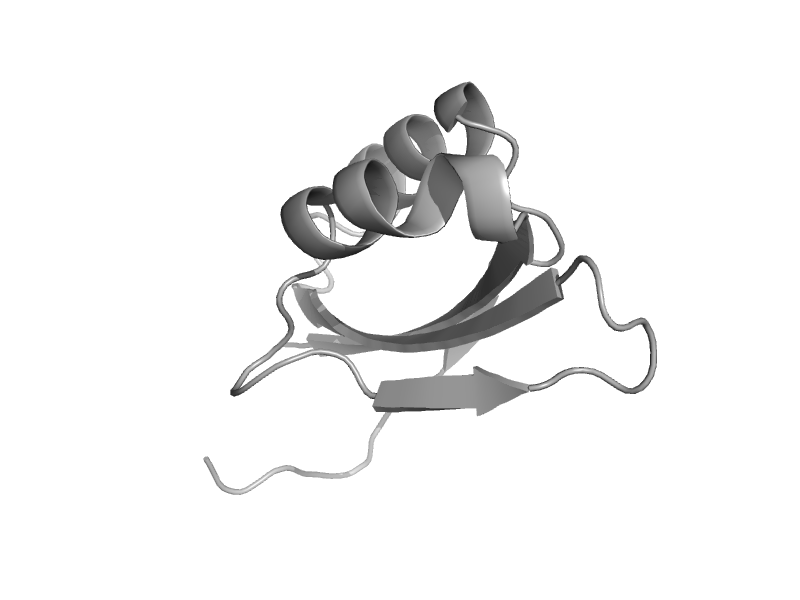 | view | |
| Cut A1 | Divalent ion tolerance proteins CutA (CutA1) | Thermus thermophilus [TaxId: 274] |  | view | |
| Hypothetical protein TM0021 | DUF190/COG1993 | Thermotoga maritima [TaxId: 2336] |  | view | |
| Cut A1 | Divalent ion tolerance proteins CutA (CutA1) | Pyrococcus horikoshii [TaxId: 53953] |  | view | |
| Hypothetical protein TTHA0516 | Prokaryotic signal transducing protein | Thermus thermophilus [TaxId: 274] |  | view | |
| automated matches | automated matches | Pyrococcus horikoshii [TaxId: 53953] |  | view | |
| Hypothetical protein RPA1041 | RPA1041-like | Rhodopseudomonas palustris [TaxId: 1076] |  | view | |
| Cut A1 | Divalent ion tolerance proteins CutA (CutA1) | Xylella fastidiosa [TaxId: 160492] |  | view | |
| automated matches | automated matches | Methanobacterium thermoautotrophicum [TaxId: 187420] |  | view | |
| automated matches | Divalent ion tolerance proteins CutA (CutA1) | Salmonella enterica [TaxId: 99287] |  | view | |
| automated matches | Prokaryotic signal transducing protein | Synechococcus elongatus [TaxId: 32046] |  | view | |
| automated matches | automated matches | Cryptosporidium parvum [TaxId: 353152] |  | view | |
| automated matches | automated matches | Ehrlichia chaffeensis [TaxId: 205920] |  | view | |
| automated matches | automated matches | Cyanobium sp. [TaxId: 180281] |  | view | |
| automated matches | automated matches | Nostoc sp. [TaxId: 103690] |  | view |
| Domain ID | Name |
|---|---|
| d4yb6a2 | 4yb6 A:225-299 |
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | Q | Y | L | M | L | D | Y | D | C | P | - | R | S | - | - | - | - | - | - | A | L | K | K | A | T | A | I | T | P | G | L | E | - | - | - | S | P | T | I | A | P | L | - | - | - | - | - | - | - | - | - | A | D | P | - | - | - | - | - | - | - | - | - | D | W | V | A | I | R | A | L | V | - | P | R | R | D | V | N | G | I | M | D | E | L | A | A | I | - | - | - | - | - | - | G | A | K | A | I | L | A | S | D | I | R | F | C | R | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | M | E | E | V | V | L | I | T | V | P | S | E | E | - | - | - | - | - | - | V | A | R | T | I | A | K | A | L | V | E | E | R | L | A | A | C | V | N | I | V | P | G | L | T | S | I | Y | R | W | Q | - | - | - | - | - | - | - | G | E | V | V | E | D | Q | E | L | L | L | L | V | K | T | - | T | T | H | A | F | P | K | L | K | E | R | V | K | A | L | H | P | Y | T | - | - | - | V | P | E | I | V | A | L | P | I | A | - | - | - | - | E | G | N | R | E | Y | L | D | W | L | R | E | N | T | G | - | - |
| - | - | - | - | - | - | - | - | - | K | L | L | K | I | Y | L | G | E | - | D | K | H | S | G | - | P | L | F | E | Y | L | V | - | R | A | Y | E | L | G | - | - | K | G | V | T | V | Y | R | G | I | G | F | G | H | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | D | L | P | I | V | L | E | I | V | D | - | E | E | E | R | I | N | L | F | L | - | - | - | - | E | I | D | I | - | - | - | - | - | F | G | L | V | F | T | A | D | V | N | V | V | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | M | I | I | V | Y | T | T | F | P | D | W | E | - | - | - | - | - | - | S | A | E | K | V | V | K | T | L | L | K | E | R | L | I | A | C | A | N | L | R | E | - | H | R | A | F | Y | W | W | E | - | - | - | - | - | - | - | G | K | I | E | E | D | K | E | V | G | A | I | L | K | T | - | R | E | D | L | W | E | E | L | K | E | R | I | K | E | L | H | P | Y | D | - | - | - | V | P | A | I | I | R | I | D | V | D | - | - | - | - | D | V | N | E | D | Y | L | K | W | L | I | E | E | T | K | K | - |
| - | - | - | - | D | L | V | P | L | K | L | V | T | I | V | A | E | - | S | L | - | - | - | - | - | - | L | E | K | R | L | V | E | E | V | K | R | L | G | - | A | K | G | Y | T | I | T | P | - | A | R | G | E | - | - | - | G | S | R | G | I | R | S | - | - | V | D | W | E | G | Q | N | I | R | L | E | T | I | V | - | S | E | E | V | A | L | R | I | L | Q | R | L | Q | E | E | Y | F | P | H | - | - | Y | A | V | I | A | Y | V | E | N | V | - | W | V | V | - | - | - | - | - | - | R | G | E | K | Y | V | - | - | - | - | - | - |
| - | V | E | V | E | H | W | N | T | L | R | L | R | I | Y | I | G | E | N | D | K | W | E | G | R | P | L | Y | K | V | I | V | E | K | L | R | E | M | G | - | I | A | G | A | T | V | Y | R | G | I | Y | G | F | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | D | L | P | I | I | V | E | V | V | D | - | R | G | H | N | I | E | K | V | V | N | V | I | K | P | M | I | K | - | - | - | - | - | D | G | M | I | T | V | E | P | T | I | V | L | W | V | G | T | Q | - | - | - | - | - | - | - | - | - | - | - | - | E | E |
| - | - | - | - | - | - | - | - | - | L | R | E | L | L | R | T | N | D | A | V | - | - | - | - | - | - | L | L | S | A | V | G | A | L | L | D | G | A | D | - | - | I | G | H | L | V | L | D | Q | - | - | - | - | - | - | - | - | N | M | S | I | L | E | G | - | - | - | - | S | L | G | V | I | P | R | R | V | L | V | - | H | E | D | D | L | A | G | A | R | R | L | L | T | D | A | G | L | A | H | E | L | R | S | D | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | S | D | V | Y | L | I | F | S | T | C | P | D | L | P | - | - | - | - | - | - | S | A | E | I | I | S | R | V | L | V | Q | E | R | L | A | A | C | V | T | Q | L | P | G | A | V | S | T | Y | R | W | Q | - | - | - | - | - | - | - | G | K | I | E | T | T | Q | E | I | Q | L | L | I | K | T | - | N | A | V | H | V | N | A | A | I | T | R | L | C | A | L | H | P | Y | R | - | - | - | L | P | E | A | I | A | V | Q | V | S | - | - | - | - | V | G | L | P | E | Y | L | T | W | I | N | T | E | I | D | - | - |
| - | - | - | - | - | - | - | G | K | R | L | V | M | L | N | I | D | - | R | K | - | - | - | - | - | - | N | L | D | R | V | R | A | L | M | P | G | M | T | - | - | - | G | P | T | V | S | E | V | - | - | - | - | - | - | - | - | - | L | S | D | - | - | - | - | - | - | - | - | N | G | V | V | A | V | H | A | V | V | - | D | E | K | E | V | F | N | L | I | N | R | L | K | A | V | - | - | - | - | - | - | G | A | R | D | I | L | V | V | P | I | E | R | I | I | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | P | A | V | V | V | L | C | T | A | P | D | E | A | - | - | - | - | - | - | T | A | Q | D | L | A | A | K | V | L | A | E | K | L | A | A | C | A | T | L | L | P | G | A | T | S | L | Y | Y | - | E | - | - | - | - | - | - | - | G | K | L | E | Q | E | Y | E | V | Q | M | I | L | K | T | - | T | V | S | H | Q | Q | A | L | I | D | C | L | K | S | H | H | P | Y | Q | - | - | - | T | P | E | L | L | V | L | P | V | T | - | - | - | - | H | G | D | T | D | Y | L | S | W | L | N | A | S | L | R | - | - |
| - | - | - | - | - | - | - | - | M | K | K | I | E | A | I | I | R | - | P | F | - | - | - | - | - | - | K | L | D | E | V | K | I | A | L | V | N | A | G | - | I | V | G | M | T | V | S | E | - | V | R | G | F | G | - | - | - | - | R | Q | K | G | S | E | Y | T | V | E | F | L | Q | K | L | K | L | E | I | V | V | - | E | D | A | Q | V | D | T | V | I | D | K | I | V | A | A | A | R | T | G | E | N | G | D | G | K | I | F | V | S | P | V | D | Q | T | I | R | I | R | T | G | E | K | N | A | D | A | I | - | - | - | - | - | - |
| T | E | T | K | I | E | S | N | I | I | L | I | Y | I | S | A | P | N | Q | D | - | - | - | - | - | - | E | A | T | S | I | A | K | T | L | V | D | E | E | L | C | A | C | V | S | I | I | P | S | V | R | S | I | Y | K | F | K | - | - | - | - | - | - | - | G | Q | V | H | D | E | N | E | V | M | L | L | V | K | T | - | T | S | Q | L | F | T | T | L | K | E | K | V | T | E | I | H | S | Y | E | - | - | - | L | P | E | I | I | A | T | K | V | V | - | - | - | - | Y | G | N | E | N | Y | I | N | W | V | N | Q | T | V | R | - | - |
| - | - | - | - | - | - | - | N | I | S | L | L | Y | T | T | T | P | T | Y | - | - | - | - | - | - | - | D | A | Y | R | I | S | N | I | L | L | E | N | K | L | I | A | C | A | N | I | F | S | - | I | T | S | V | Y | - | - | - | - | - | - | - | - | - | - | - | D | I | H | N | N | T | E | C | A | I | I | L | K | T | - | T | N | D | L | V | Q | H | A | T | N | K | I | - | A | I | H | P | Y | D | - | - | - | T | P | A | I | I | T | I | D | P | T | - | - | - | - | N | A | N | - | D | F | I | Q | W | V | N | D | C | T | A | L | - |
| - | - | - | - | - | S | Q | Q | V | W | K | L | V | I | I | T | E | - | E | I | - | - | - | - | - | - | L | L | K | K | V | S | K | I | I | K | E | A | G | - | A | S | G | Y | T | V | L | A | - | A | A | G | E | G | S | R | N | V | R | S | T | G | E | P | S | V | S | H | A | Y | S | N | I | K | F | E | V | L | T | A | S | R | E | L | A | D | Q | I | Q | D | K | V | V | A | K | Y | F | D | D | - | - | Y | S | C | I | T | Y | I | S | T | V | - | E | A | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | M | K | I | A | L | T | N | L | P | - | P | E | - | - | - | - | - | - | H | G | E | R | I | A | R | L | L | V | E | E | H | I | V | A | C | V | N | L | Y | P | - | V | H | S | I | Y | S | W | K | - | - | - | - | - | - | - | G | E | V | C | S | E | A | E | V | T | L | M | M | K | V | - | S | T | Q | G | I | E | R | L | K | Q | R | I | C | E | L | H | P | Y | E | - | - | - | L | P | E | F | V | V | I | E | V | D | N | - | - | N | A | S | L | R | E | Y | I | D | F | V | K | G | E | T | H | L | Y |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1nh8a2 | A | P9WMN1 | GO:0000105 |
| d1nh8a2 | A | P9WMN1 | GO:0000287 |
| d1nh8a2 | A | P9WMN1 | GO:0003879 |
| d1nh8a2 | A | P9WMN1 | GO:0005515 |
| d1nh8a2 | A | P9WMN1 | GO:0005524 |
| d1nh8a2 | A | P9WMN1 | GO:0005737 |
| d1nh8a2 | A | P9WMN1 | GO:0005886 |
| d1nh8a2 | A | P9WMN1 | GO:0016208 |
| d1nh8a2 | A | P9WMN1 | GO:0016757 |
| d1nh8a2 | A | P9WMN1 | GO:0046872 |
| d1nzaa_ | A | Q7SIA8 | GO:0005507 |
| d1nzaa_ | A | Q7SIA8 | GO:0005737 |
| d1nzaa_ | A | Q7SIA8 | GO:0010038 |
| d1ukua_ | A | O58720 | GO:0005507 |
| d1ukua_ | A | O58720 | GO:0005737 |
| d1ukua_ | A | O58720 | GO:0010038 |
| d1ukua_ | A | O58720 | GO:0046872 |
| d2cz4a1 | A | Q5SKX7 | GO:0006808 |
| d2cz4a1 | A | Q5SKX7 | GO:0030234 |
