solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1eeja2 ( 1 ) ddaaIqqtLakmgikssdiqpApvagmktVlTns----gvlyItddGkhi
d1t3ba2 ( 2 ) daaIlis---------viSp---sgiktAvTq-----giLyVeg---yl
d1v58a2 ( 2 ) elpapVkaiekqgitiiktfdAp-ggmkGYlGkyqdmgvtIyltpdgkhA
60
d1eeja2 ( 47 ) iqgpmydvsgtapv
d1t3ba2 ( 47 ) fegklye--lngpv
d1v58a2 ( 51 ) isgymynek---ge
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Disulfide bond isomerase, DsbC, N-terminal domain | DsbC/DsbG N-terminal domain-like | Escherichia coli [TaxId: 562] | 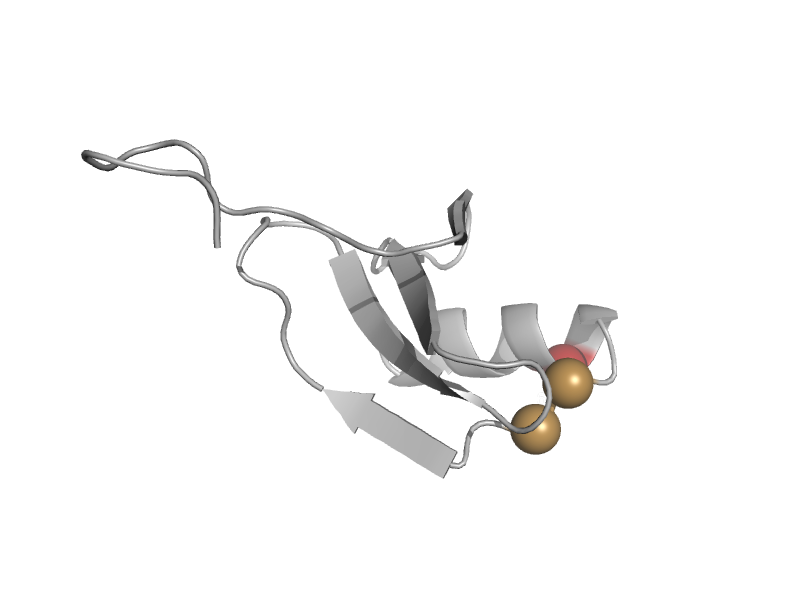 | view | |
| Disulfide bond isomerase, DsbC, N-terminal domain | DsbC/DsbG N-terminal domain-like | Haemophilus influenzae [TaxId: 727] | 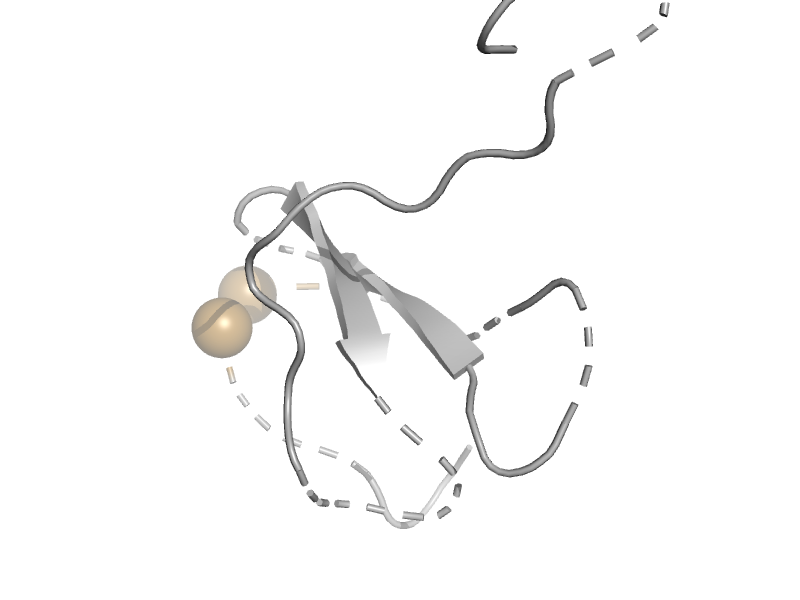 | view | |
| Thiol:disulfide interchange protein DsbG, N-terminal domain | DsbC/DsbG N-terminal domain-like | Escherichia coli [TaxId: 562] | 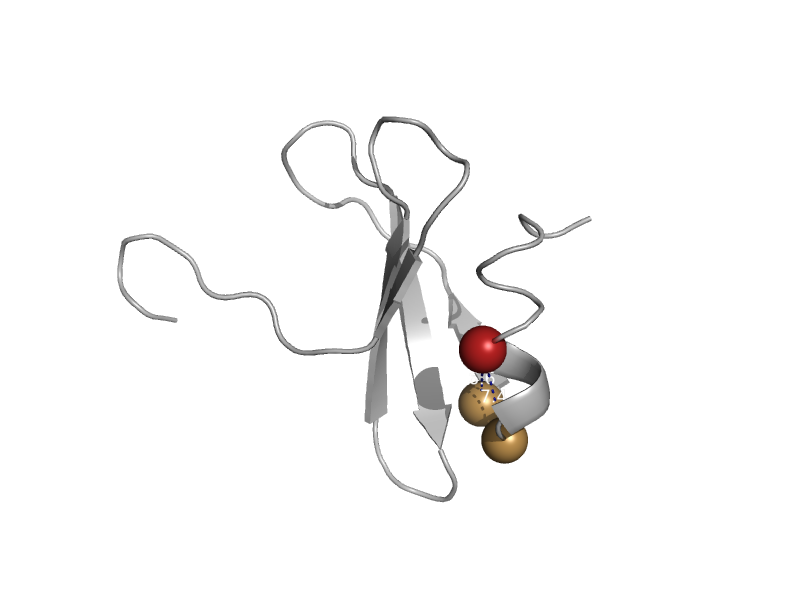 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D | D | A | A | I | Q | Q | T | L | A | K | M | G | I | K | S | S | D | I | Q | P | A | P | V | A | G | M | K | T | V | L | T | N | S | - | - | - | - | G | V | L | Y | I | T | D | D | G | K | H | I | I | Q | G | P | M | Y | D | V | S | G | T | A | P | V |
| - | D | A | A | I | L | I | S | - | - | - | - | - | - | - | - | - | V | I | S | P | - | - | - | S | G | I | K | T | A | V | T | Q | - | - | - | - | - | G | I | L | Y | V | E | G | - | - | - | Y | L | F | E | G | K | L | Y | E | - | - | L | N | G | P | V |
| E | L | P | A | P | V | K | A | I | E | K | Q | G | I | T | I | I | K | T | F | D | A | P | - | G | G | M | K | G | Y | L | G | K | Y | Q | D | M | G | V | T | I | Y | L | T | P | D | G | K | H | A | I | S | G | Y | M | Y | N | E | K | - | - | - | G | E |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1eeja2 | A | P0AEG6 | GO:0003756 |
| d1eeja2 | A | P0AEG6 | GO:0015035 |
| d1eeja2 | A | P0AEG6 | GO:0015036 |
| d1eeja2 | A | P0AEG6 | GO:0030288 |
| d1eeja2 | A | P0AEG6 | GO:0042597 |
| d1eeja2 | A | P0AEG6 | GO:0042803 |
| d1eeja2 | A | P0AEG6 | GO:0046688 |
| d1eeja2 | A | P0AEG6 | GO:0061077 |
| d1t3ba2 | A | P45111 | GO:0042597 |
| d1v58a2 | A | P77202 | GO:0003756 |
| d1v58a2 | A | P77202 | GO:0030288 |
| d1v58a2 | A | P77202 | GO:0042597 |
| d1v58a2 | A | P77202 | GO:0042803 |
| d1v58a2 | A | P77202 | GO:0061077 |
