solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1x9ya2 ( 41 ) kqLeInVksdkvPqkVkdlAq
d2bk7b1 ( 1 ) mei-------ppt-nypAs
d2bk7b2 ( 99 ) pdeedntfYqrLksmkepleaqnIpdnfgnvspe-mtlVL
d2ch9a1 ( 25 ) tcsqdlnsrv----------------kpgfpkti-------ktn-dpgVl
d2gu3a1 ( 99 ) gised
d2gu3a2 ( 34 ) keeghe
d2octa- ( 2 ) ms-gapsatqpataetqhiad
d3mwza1 ( 2 ) elalrggyrersnqddpeyl
d4eyca1 ( 31 ) qvlsyk
d4it7a- ( 6 ) ggfstk-------dvn-dpkIq
d4n6ob1 ( 11 ) gelrdl-------spd-dpqVq
d5o46a- ( 1 ) f---pgvwrkhhpdvdprYk
d5zc1a- ( 36 ) vmpi----------------cggisaar-------iPt-adekk
d6uioa- ( 31 ) aqnyfgsi-------nis-nanvk
d6ztka- ( 13 ) gwktqdpt-npkfe
aaaa
60 70 80 90 100
d1x9ya2 ( 62 ) qqfagyAkalDkqsnaktgkyeLGeAFKIYkfnGeednSYyyPVIkdgkI
d2bk7b1 ( 12 ) rAAlvAqnyInyqqGtphrvFevqkVkqAsmedip----------grghk
d2bk7b2 ( 138 ) hLAwvACgyIiwqnstedtwykMvkIqtVkQvqrn----------ddfIE
d2ch9a1 ( 51 ) qAArySVekfnnctndm-flFkesrItrAlvqiv------------kGlk
d2gu3a1 ( 104 ) kAakiIkdeg-----------lvskqkeVhlare-----------gnvlL
d2gu3a2 ( 40 ) aAaaeAkket-----------dLahvdqvetfvg------------keky
d2octa- ( 22 ) q-vrsqleeky---nkkfpvfkavsfks-------------qvv----ag
d3mwza1 ( 22 ) e-AhyATstwSaqqpgkthfDTVveV-kVeTQtv------------agtN
d4eyca1 ( 37 ) eAVlrAIdgInqrssd-anlYrlldldprtmd--g----------dpdtp
d4it7a- ( 20 ) aLAgkAlqrInaasndl-fqQtivkVisAktqvv------------agtn
d4n6ob1 ( 25 ) kAAqaAVasyNmgsnsi-yYfrdthiikAqsqlv------------aGik
d5o46a- ( 18 ) eWAhfAIssqv---enrtnfDTlmtLisVesqvi------------agvD
d5zc1a- ( 56 ) kLepvLlqsLyahlg---skPtsAeVvlVAtqvv------------agtN
d6uioa- ( 47 ) qAvwfAMkeynkesedk-yvFlvdkilhAklqit------------drme
d6ztka- ( 26 ) nlAhyAVstqv---egreyydTVleLleVqtqiv------------aGvn
aaaaaaaaa a bbbbbbbbbbbb bb
110 120 130 140 150
d1x9ya2 ( 112 ) vyTLtLsPkn---kdDlnkskedmnYsVkisnfIAkdLdqikdknsnITV
d2bk7b1 ( 52 ) yrLkFaVeEiiqk--------------------------------qvkvn
d2bk7b2 ( 178 ) LdYtIlLhniasq---------------------------------eiip
d2ch9a1 ( 88 ) YmLeVeIgrTtçkkn----q----------hlrLddÇdfQtnhtlkqtls
d2gu3a1 ( 132 ) weVTYldke--------------------------------------gqy
d2gu3a2 ( 67 ) yvVkGtdkk--------------------------------------gta
d2octa- ( 51 ) tnyfikvhvg---d-------------------------------edfvH
d3mwza1 ( 60 ) yrLtLkVAeStçelt--s------------tynkdtÇqAna---naaqrt
d4eyca1 ( 75 ) KpVSFtVkeTvÇprttq--------------qspedÇdfkk---dGlvkr
d4it7a- ( 57 ) TvLeLlIApTsçrknetsag---------------nçeavs---nGtkqi
d4n6ob1 ( 62 ) YfLtMeMgSTdcrktrvtgd----------hvdlttCplAag-aqqeklr
d5o46a- ( 53 ) ykLkMkVAeStÇvigv-d------------syskerÇylkv---nvpyMl
d5zc1a- ( 91 ) YFAKVkVnn--------------------------------------dhy
d6uioa- ( 84 ) YqidvqisrSnÇkkplnnte---------------nÇipqkkpelekkms
d6ztka- ( 61 ) ykLkFtTTqStçkiesgv------------eYskelÇqpkt---nkveav
bbbbbbbb bb
160 170 180 190 200
d1x9ya2 ( 159 ) LtdekGfYFeed--g--kvrlVkatp--lanNikekesaktVspqLkqel
d2bk7b1 ( 70 ) CtA---eVlypstg---qetapeVnftfe--getgkn
d2bk7b2 ( 195 ) wqM---qVlwhPq---ygtkVkhnsrlpk
d2ch9a1 ( 124 ) ÇyS---eVwvVpwl--qhfeVpvlrçh
d2gu3a1 ( 144 ) SlS---yVdft---tG---kilknitp
d2gu3a2 ( 79 ) lyV---wVPad---kk---akilskeake
d2octa- ( 67 ) lrv---fqslph--enksltlsnyqtnkak------------------hd
d3mwza1 ( 93 ) ÇtT---viyrnq----geksinsFeçaaa
d4eyca1 ( 108 ) Çmg---tVtlnqarg-----sfdIsÇdkdnk
d4it7a- ( 89 ) ÇtV---aIwekpwe--nfeeItikeçksa
d4n6ob1 ( 101 ) ÇdF---eVlvvpwq--nssqLlkhnçvqm
d5o46a- ( 87 ) ÇtA---vVnympwe--hktilksYdçsdrvygv
d5zc1a- ( 103 ) IHT---rVyeqlpcyggaleLhsVqmnktdtdpLdyf
d6uioa- ( 119 ) ÇsF---lvGAlPwn--geFnllskeçkdv
d6ztka- ( 96 ) ÇtS---iIytvpwq--nikrvlsYhçdap
bbb bbbb bbbbbbbb
d1x9ya2 ( 203 ) kttVtPtkv
d2bk7b1
d2bk7b2
d2ch9a1
d2gu3a1
d2gu3a2
d2octa- ( 94 ) eltyf
d3mwza1
d4eyca1
d4it7a-
d4n6ob1
d5o46a-
d5zc1a-
d6uioa-
d6ztka-
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Staphopain B, prodomain | Staphopain B, prodomain | Staphylococcus aureus [TaxId: 1280] | 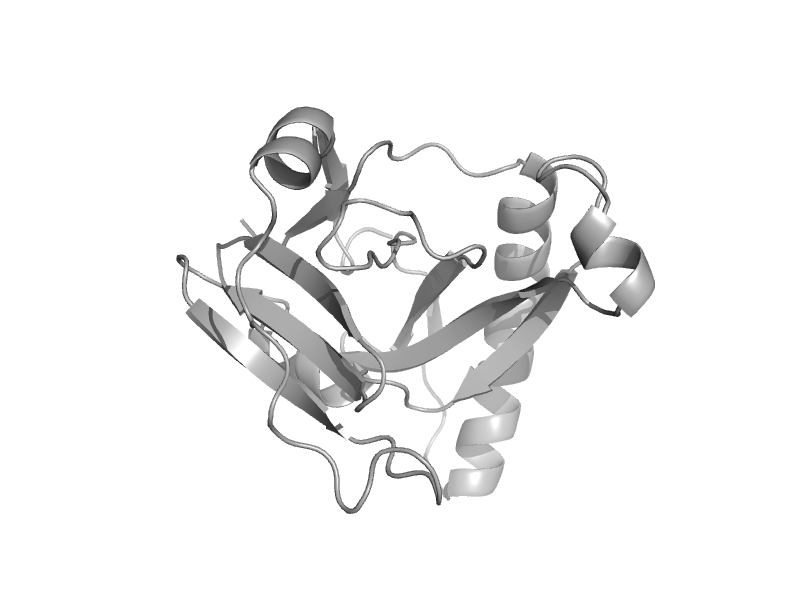 | view | |
| Latexin | Latexin-like | Human (Homo sapiens) [TaxId: 9606] | 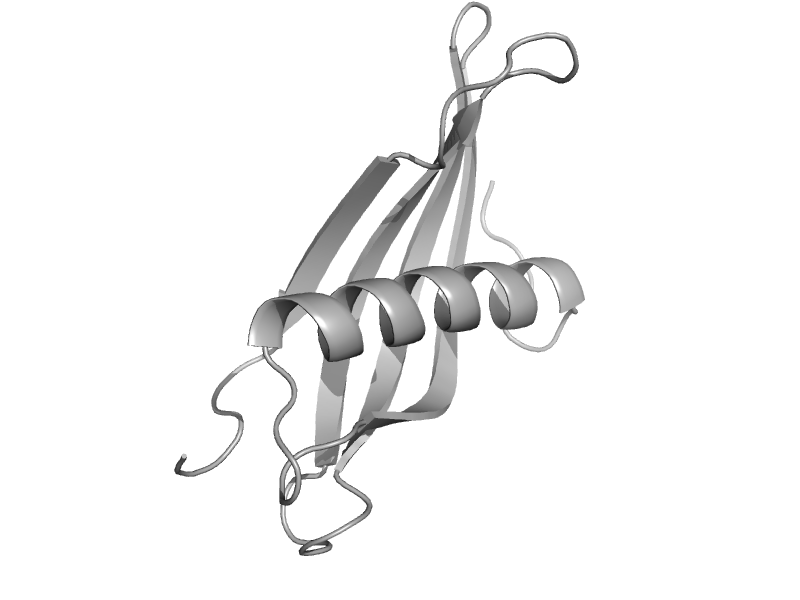 | view | |
| Latexin | Latexin-like | Human (Homo sapiens) [TaxId: 9606] | 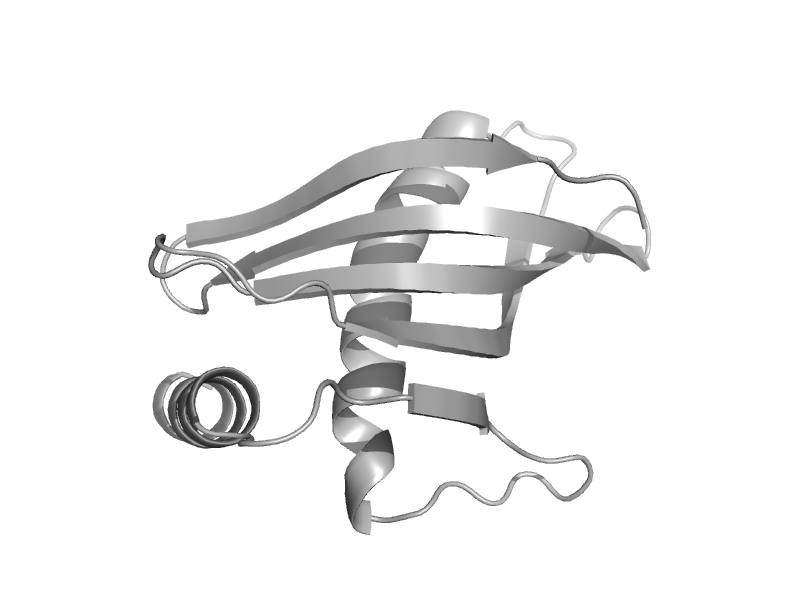 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 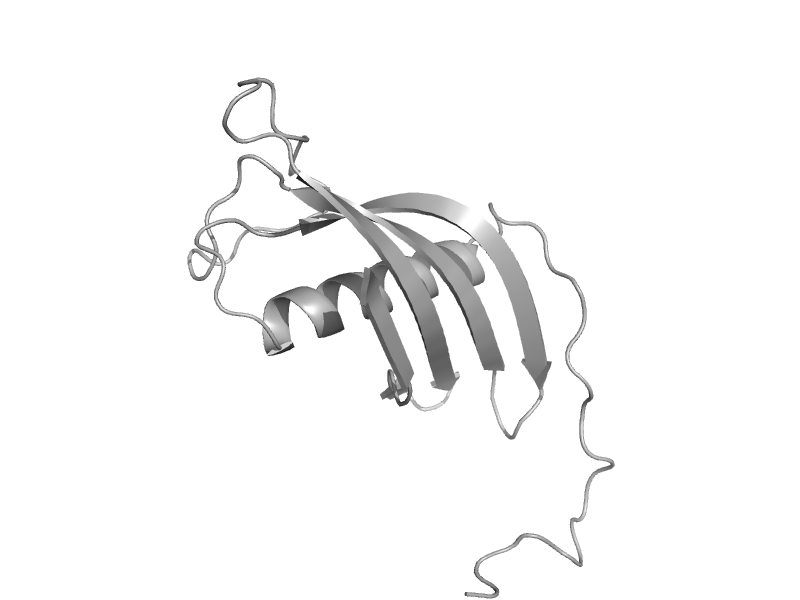 | view | |
| Uncharacterized protein YpmB | PepSY-like | Bacillus subtilis [TaxId: 1423] |  | view | |
| Uncharacterized protein YpmB | PepSY-like | Bacillus subtilis [TaxId: 1423] |  | view | |
| automated matches | Cystatins | Human (Homo sapiens) [TaxId: 9606] |  | view | |
| automated matches | automated matches | Ixodes scapularis [TaxId: 6945] |  | view | |
| automated matches | Cathelicidin motif | Human (Homo sapiens) [TaxId: 9606] |  | view | |
| automated matches | automated matches | Ascaris lumbricoides [TaxId: 6252] |  | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] |  | view | |
| automated matches | automated matches | Ixodes ricinus [TaxId: 34613] |  | view | |
| automated matches | automated matches | Clonorchis sinensis [TaxId: 79923] |  | view | |
| automated matches | automated matches | Mouse (Mus musculus) [TaxId: 10090] |  | view | |
| automated matches | automated matches | Ixodes ricinus [TaxId: 34613] |  | view |
| Domain ID | Name |
|---|---|
| d6vlpa- | 6vlp A: |
| d2o9ux- | 2o9u X: |
| d3nx0a- | 3nx0 A: |
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | Q | L | E | I | N | V | K | S | D | K | V | P | Q | K | V | K | D | L | A | Q | Q | Q | F | A | G | Y | A | K | A | L | D | K | Q | S | N | A | K | T | G | K | Y | E | L | G | E | A | F | K | I | Y | K | F | N | G | E | E | D | N | S | Y | Y | Y | P | V | I | K | D | G | K | I | V | Y | T | L | T | L | S | P | K | N | - | - | - | K | D | D | L | N | K | S | K | E | D | M | N | Y | S | V | K | I | S | N | F | I | A | K | D | L | D | Q | I | K | D | K | N | S | N | I | T | V | L | T | D | E | K | G | F | Y | F | E | E | D | - | - | G | - | - | K | V | R | L | V | K | A | T | P | - | - | L | A | N | N | I | K | E | K | E | S | A | K | T | V | S | P | Q | L | K | Q | E | L | K | T | T | V | T | P | T | K | V |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | E | I | - | - | - | - | - | - | - | P | P | T | - | N | Y | P | A | S | R | A | A | L | V | A | Q | N | Y | I | N | Y | Q | Q | G | T | P | H | R | V | F | E | V | Q | K | V | K | Q | A | S | M | E | D | I | P | - | - | - | - | - | - | - | - | - | - | G | R | G | H | K | Y | R | L | K | F | A | V | E | E | I | I | Q | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | V | K | V | N | C | T | A | - | - | - | E | V | L | Y | P | S | T | G | - | - | - | Q | E | T | A | P | E | V | N | F | T | F | E | - | - | G | E | T | G | K | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | P | D | E | E | D | N | T | F | Y | Q | R | L | K | S | M | K | E | P | L | E | A | Q | N | I | P | D | N | F | G | N | V | S | P | E | - | M | T | L | V | L | H | L | A | W | V | A | C | G | Y | I | I | W | Q | N | S | T | E | D | T | W | Y | K | M | V | K | I | Q | T | V | K | Q | V | Q | R | N | - | - | - | - | - | - | - | - | - | - | D | D | F | I | E | L | D | Y | T | I | L | L | H | N | I | A | S | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | I | I | P | W | Q | M | - | - | - | Q | V | L | W | H | P | Q | - | - | - | Y | G | T | K | V | K | H | N | S | R | L | P | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| T | C | S | Q | D | L | N | S | R | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | P | G | F | P | K | T | I | - | - | - | - | - | - | - | K | T | N | - | D | P | G | V | L | Q | A | A | R | Y | S | V | E | K | F | N | N | C | T | N | D | M | - | F | L | F | K | E | S | R | I | T | R | A | L | V | Q | I | V | - | - | - | - | - | - | - | - | - | - | - | - | K | G | L | K | Y | M | L | E | V | E | I | G | R | T | T | C | K | K | N | - | - | - | - | Q | - | - | - | - | - | - | - | - | - | - | H | L | R | L | D | D | C | D | F | Q | T | N | H | T | L | K | Q | T | L | S | C | Y | S | - | - | - | E | V | W | V | V | P | W | L | - | - | Q | H | F | E | V | P | V | L | R | C | H | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | I | S | E | D | K | A | A | K | I | I | K | D | E | G | - | - | - | - | - | - | - | - | - | - | - | L | V | S | K | Q | K | E | V | H | L | A | R | E | - | - | - | - | - | - | - | - | - | - | - | G | N | V | L | L | W | E | V | T | Y | L | D | K | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | Q | Y | S | L | S | - | - | - | Y | V | D | F | T | - | - | - | T | G | - | - | - | K | I | L | K | N | I | T | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | E | E | G | H | E | A | A | A | A | E | A | K | K | E | T | - | - | - | - | - | - | - | - | - | - | - | D | L | A | H | V | D | Q | V | E | T | F | V | G | - | - | - | - | - | - | - | - | - | - | - | - | K | E | K | Y | Y | V | V | K | G | T | D | K | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | T | A | L | Y | V | - | - | - | W | V | P | A | D | - | - | - | K | K | - | - | - | A | K | I | L | S | K | E | A | K | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | S | - | G | A | P | S | A | T | Q | P | A | T | A | E | T | Q | H | I | A | D | Q | - | V | R | S | Q | L | E | E | K | Y | - | - | - | N | K | K | F | P | V | F | K | A | V | S | F | K | S | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | V | V | - | - | - | - | A | G | T | N | Y | F | I | K | V | H | V | G | - | - | - | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | D | F | V | H | L | R | V | - | - | - | F | Q | S | L | P | H | - | - | E | N | K | S | L | T | L | S | N | Y | Q | T | N | K | A | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | H | D | E | L | T | Y | F | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | L | A | L | R | G | G | Y | R | E | R | S | N | Q | D | D | P | E | Y | L | E | - | A | H | Y | A | T | S | T | W | S | A | Q | Q | P | G | K | T | H | F | D | T | V | V | E | V | - | K | V | E | T | Q | T | V | - | - | - | - | - | - | - | - | - | - | - | - | A | G | T | N | Y | R | L | T | L | K | V | A | E | S | T | C | E | L | T | - | - | S | - | - | - | - | - | - | - | - | - | - | - | - | T | Y | N | K | D | T | C | Q | A | N | A | - | - | - | N | A | A | Q | R | T | C | T | T | - | - | - | V | I | Y | R | N | Q | - | - | - | - | G | E | K | S | I | N | S | F | E | C | A | A | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | V | L | S | Y | K | E | A | V | L | R | A | I | D | G | I | N | Q | R | S | S | D | - | A | N | L | Y | R | L | L | D | L | D | P | R | T | M | D | - | - | G | - | - | - | - | - | - | - | - | - | - | D | P | D | T | P | K | P | V | S | F | T | V | K | E | T | V | C | P | R | T | T | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | S | P | E | D | C | D | F | K | K | - | - | - | D | G | L | V | K | R | C | M | G | - | - | - | T | V | T | L | N | Q | A | R | G | - | - | - | - | - | S | F | D | I | S | C | D | K | D | N | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | G | F | S | T | K | - | - | - | - | - | - | - | D | V | N | - | D | P | K | I | Q | A | L | A | G | K | A | L | Q | R | I | N | A | A | S | N | D | L | - | F | Q | Q | T | I | V | K | V | I | S | A | K | T | Q | V | V | - | - | - | - | - | - | - | - | - | - | - | - | A | G | T | N | T | V | L | E | L | L | I | A | P | T | S | C | R | K | N | E | T | S | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | C | E | A | V | S | - | - | - | N | G | T | K | Q | I | C | T | V | - | - | - | A | I | W | E | K | P | W | E | - | - | N | F | E | E | I | T | I | K | E | C | K | S | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | E | L | R | D | L | - | - | - | - | - | - | - | S | P | D | - | D | P | Q | V | Q | K | A | A | Q | A | A | V | A | S | Y | N | M | G | S | N | S | I | - | Y | Y | F | R | D | T | H | I | I | K | A | Q | S | Q | L | V | - | - | - | - | - | - | - | - | - | - | - | - | A | G | I | K | Y | F | L | T | M | E | M | G | S | T | D | C | R | K | T | R | V | T | G | D | - | - | - | - | - | - | - | - | - | - | H | V | D | L | T | T | C | P | L | A | A | G | - | A | Q | Q | E | K | L | R | C | D | F | - | - | - | E | V | L | V | V | P | W | Q | - | - | N | S | S | Q | L | L | K | H | N | C | V | Q | M | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | F | - | - | - | P | G | V | W | R | K | H | H | P | D | V | D | P | R | Y | K | E | W | A | H | F | A | I | S | S | Q | V | - | - | - | E | N | R | T | N | F | D | T | L | M | T | L | I | S | V | E | S | Q | V | I | - | - | - | - | - | - | - | - | - | - | - | - | A | G | V | D | Y | K | L | K | M | K | V | A | E | S | T | C | V | I | G | V | - | D | - | - | - | - | - | - | - | - | - | - | - | - | S | Y | S | K | E | R | C | Y | L | K | V | - | - | - | N | V | P | Y | M | L | C | T | A | - | - | - | V | V | N | Y | M | P | W | E | - | - | H | K | T | I | L | K | S | Y | D | C | S | D | R | V | Y | G | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | V | M | P | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | G | G | I | S | A | A | R | - | - | - | - | - | - | - | I | P | T | - | A | D | E | K | K | K | L | E | P | V | L | L | Q | S | L | Y | A | H | L | G | - | - | - | S | K | P | T | S | A | E | V | V | L | V | A | T | Q | V | V | - | - | - | - | - | - | - | - | - | - | - | - | A | G | T | N | Y | F | A | K | V | K | V | N | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | H | Y | I | H | T | - | - | - | R | V | Y | E | Q | L | P | C | Y | G | G | A | L | E | L | H | S | V | Q | M | N | K | T | D | T | D | P | L | D | Y | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | Q | N | Y | F | G | S | I | - | - | - | - | - | - | - | N | I | S | - | N | A | N | V | K | Q | A | V | W | F | A | M | K | E | Y | N | K | E | S | E | D | K | - | Y | V | F | L | V | D | K | I | L | H | A | K | L | Q | I | T | - | - | - | - | - | - | - | - | - | - | - | - | D | R | M | E | Y | Q | I | D | V | Q | I | S | R | S | N | C | K | K | P | L | N | N | T | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | C | I | P | Q | K | K | P | E | L | E | K | K | M | S | C | S | F | - | - | - | L | V | G | A | L | P | W | N | - | - | G | E | F | N | L | L | S | K | E | C | K | D | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | W | K | T | Q | D | P | T | - | N | P | K | F | E | N | L | A | H | Y | A | V | S | T | Q | V | - | - | - | E | G | R | E | Y | Y | D | T | V | L | E | L | L | E | V | Q | T | Q | I | V | - | - | - | - | - | - | - | - | - | - | - | - | A | G | V | N | Y | K | L | K | F | T | T | T | Q | S | T | C | K | I | E | S | G | V | - | - | - | - | - | - | - | - | - | - | - | - | E | Y | S | K | E | L | C | Q | P | K | T | - | - | - | N | K | V | E | A | V | C | T | S | - | - | - | I | I | Y | T | V | P | W | Q | - | - | N | I | K | R | V | L | S | Y | H | C | D | A | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1x9ya2 | A | P0C1S6 | GO:0005576 |
| d1x9ya2 | A | P0C1S6 | GO:0006508 |
| d1x9ya2 | A | P0C1S6 | GO:0008234 |
