solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1fm0d- ( 1 ) mIkVlFfaqvrelVg---tdatevaad---fptVeaLrqhMAaqs
d1ryja- ( 4 ) mvigMkFtvit---dd-------gkkilesg--aprrikdvlgel----
d1tygb- ( 1 ) mLqlng------------kdvkwk--dtgtIqdLLasy----
d1v8ca1 ( 1 ) PkVnLyatFrdlTg---ksqlelpga-----tVgeVLenLVray
d1vjka1 ( 2 ) vkVkVYfarFrqlAg---vdeeeIeLp--egArVrdLIeeIkkrh
d1zud2- ( 2 ) qIlFnd------------qamqca--agqtVheLLeql----
d2cu3a1 ( 2 ) VwlngeprplegktLkeVLee-----
d2hj1a1 ( 11 ) nqinieiayafp-------eryylksfqVde--gitvqtaitqSgIls
d2k9xa- ( 1 ) msnhnhitvqFaggCellfa--kqtslqldgvvptgtnlnglvqllKtny
d2qjla- ( 1 ) mVnVkVeFlggLdaIFg--kqrvhkIkmdkedpVtVgdLIdhIvstm
d4idia- ( 2 ) kvkIeLkFlggLesyLedksknyvtLeIds-keLnFeNLIAFIrdnI
bbbb bbbb aaaaaaaa
60 70 80 90 100
d1fm0d- ( 40 ) d--------------------rwalaLedg----------kLlAaVnqtl
d1ryja- ( 37 ) -----------------------------eipi------etvvvkKngqi
d1tygb- ( 26 ) -----------------------------qlen------kiviVernkei
d1v8ca1 ( 37 ) ---paLkeeLfeg----------------------eglaerVsVflegrd
d1vjka1 ( 43 ) e--------------------kFeevFgegydd------AdVNIavngry
d1zud2- ( 26 ) -----------------------------dqrq------agAaLainqqi
d2cu3a1 ( 24 ) -----------------------------------gvelkgvaVlLneea
d2hj1a1 ( 50 ) qfpeidlstnkigifsrpikltdvlkegdrieiyrpll
d2k9xa- ( 49 ) vker--pdllvdq--------------------tgqtlrpGilvlvnscd
d2qjla- ( 46 ) InnpndvsiFied----------------------dsIrpgIiTlIndtd
d4idia- ( 48 ) Iekk---fvFSdyddeklckVvdnkeysnYnLkdkAkikpgIiVlVneyd
bbbb bb
110 120
d1fm0d- ( 60 ) v-sfd-----hpLtdgdeVaFfppvtgg
d1ryja- ( 52 ) v-ide-----eeifdgdiievirviygg
d1tygb- ( 41 ) I-gke-ryheveLcdrdvIeivhfvgg
d1v8ca1 ( 62 ) --VrylqglsTpLspgatLdLfPpvagg
d1vjka1 ( 69 ) v-swd-----eeL-dgdvVgVfppv
d1zud2- ( 41 ) v-pre-qWaqhiVqdgdqIlLfqviagg
d2cu3a1 ( 39 ) flGle--VpdrpLrdgdvVevvalq--g
d2hj1a1
d2k9xa- ( 77 ) --AevvggmdyvlndgDtveFistlhgg
d2qjla- ( 74 ) --welegekdyiLedgdiIsFtstlhGg
d4idia- ( 100 ) --weilgtysyqIkndd-IcFlst
bbbbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Molybdopterin synthase subunit MoaD | MoaD | Escherichia coli [TaxId: 562] | 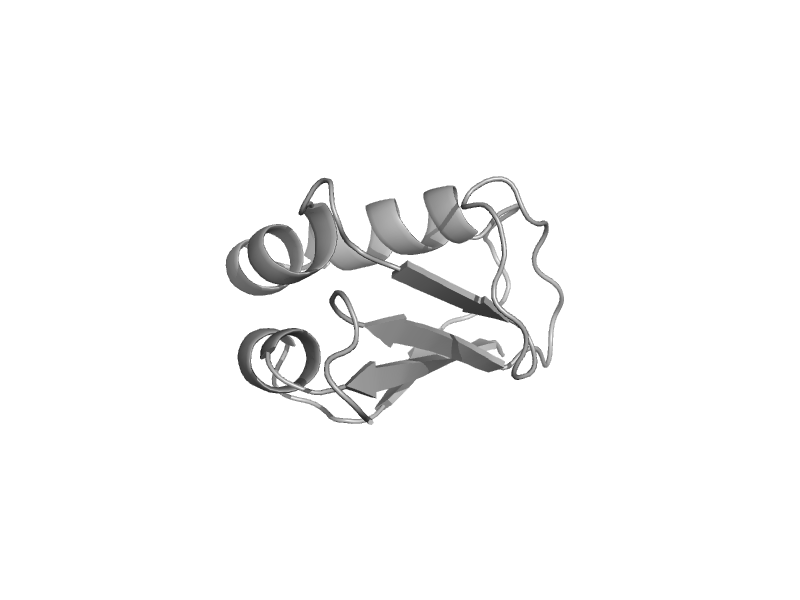 | view | |
| Hypothetical protein MTH1743 | ThiS | Methanobacterium thermoautotrophicum [TaxId: 145262] | 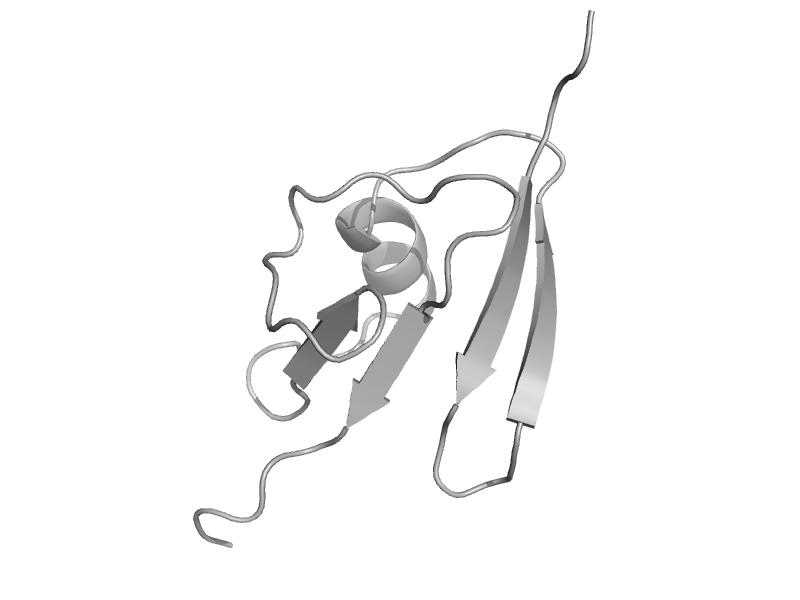 | view | |
| Thiamin biosynthesis sulfur carrier protein ThiS | ThiS | Bacillus subtilis [TaxId: 1423] | 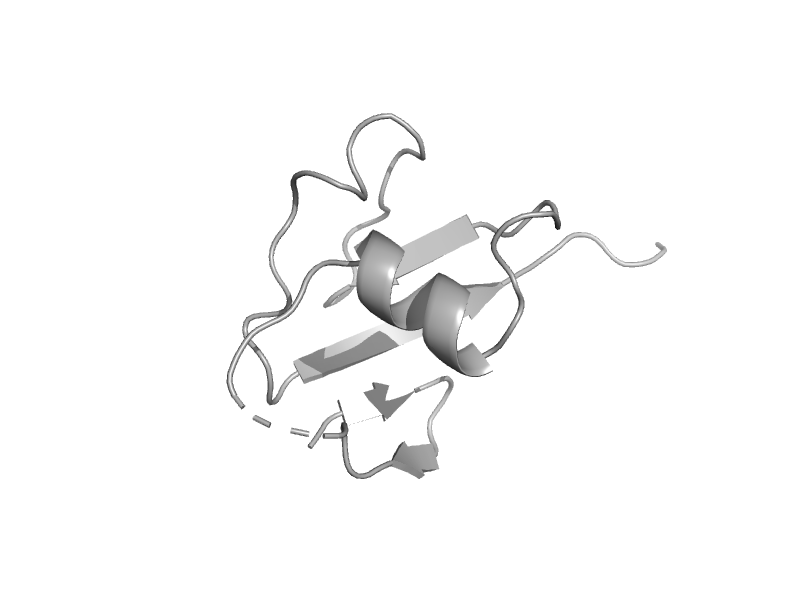 | view | |
| MoaD-related protein, N-terminal domain | MoaD | Thermus thermophilus [TaxId: 274] | 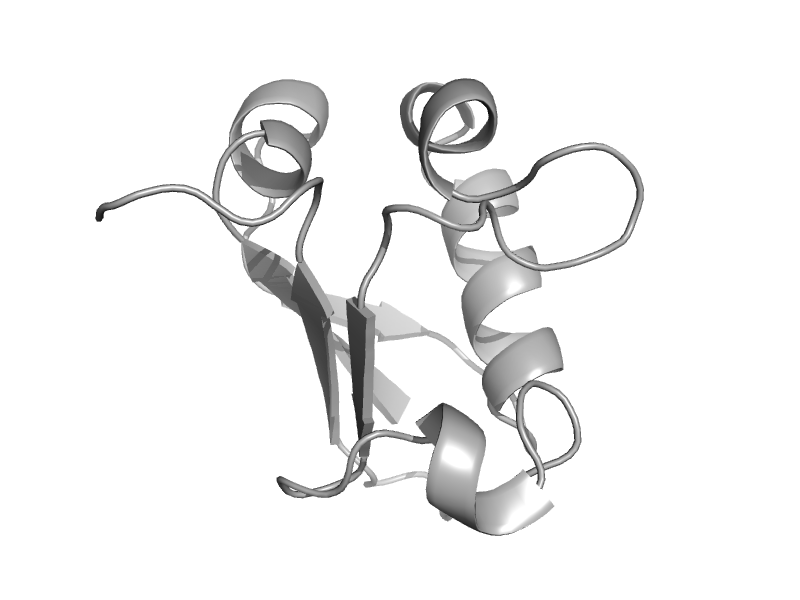 | view | |
| Molybdopterin synthase subunit MoaD | MoaD | Pyrococcus furiosus [TaxId: 2261] | 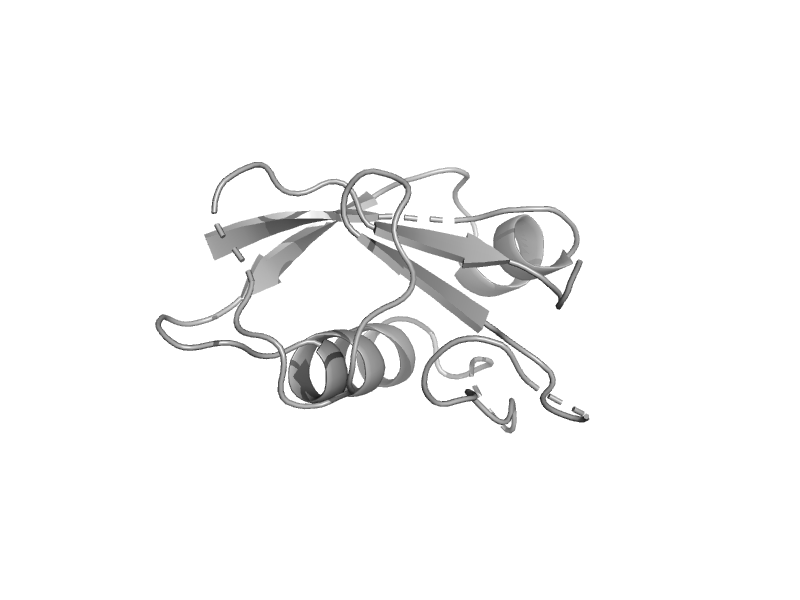 | view | |
| Thiamin biosynthesis sulfur carrier protein ThiS | ThiS | Escherichia coli [TaxId: 562] | 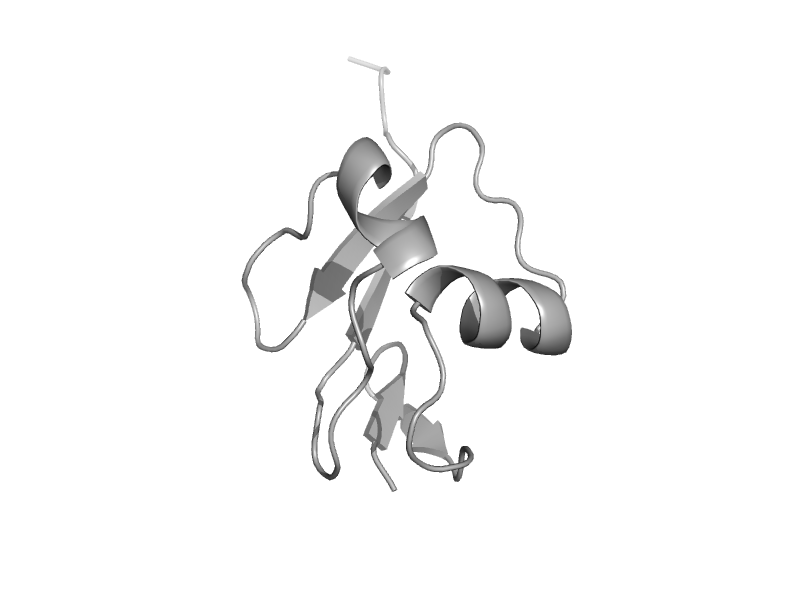 | view | |
| Uncharacterised protein TTHA0675 | ThiS | Thermus thermophilus [TaxId: 274] | 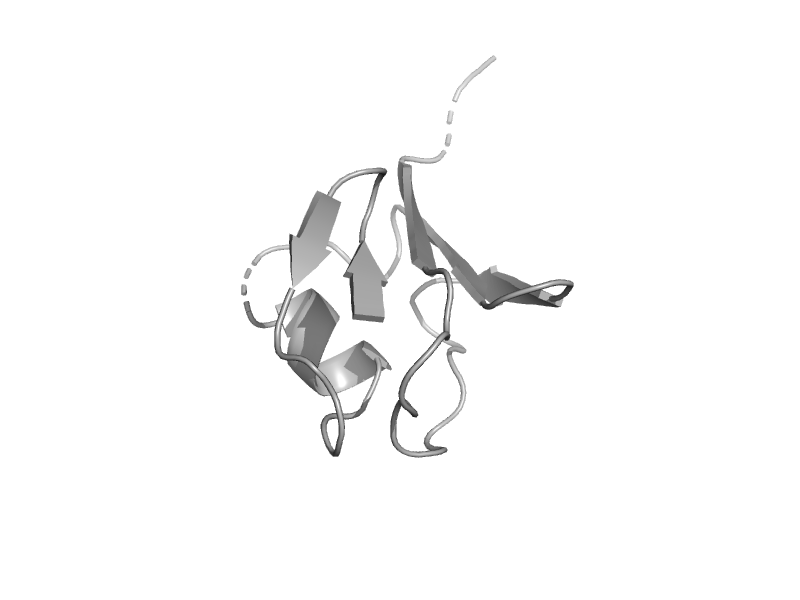 | view | |
| Hypothetical protein HI0395 | HI0395-like | Haemophilus influenzae [TaxId: 727] | 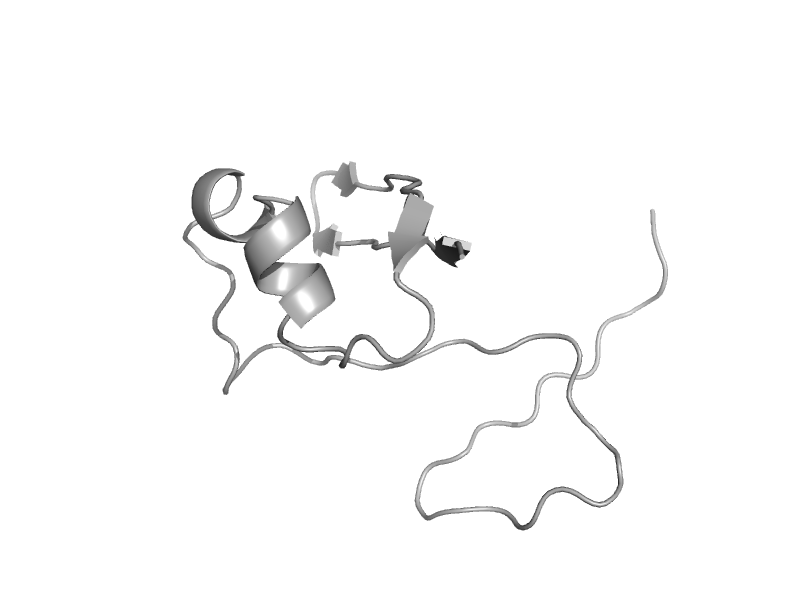 | view | |
| automated matches | automated matches | Trypanosome (Trypanosoma brucei) [TaxId: 5691] |  | view | |
| automated matches | automated matches | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] |  | view | |
| automated matches | automated matches | Plasmodium yoelii [TaxId: 73239] |  | view |
| Domain ID | Name |
|---|---|
| d5ldab- | 5lda B: |
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | M | I | K | V | L | F | F | A | Q | V | R | E | L | V | G | - | - | - | T | D | A | T | E | V | A | A | D | - | - | - | F | P | T | V | E | A | L | R | Q | H | M | A | A | Q | S | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | W | A | L | A | L | E | D | G | - | - | - | - | - | - | - | - | - | - | K | L | L | A | A | V | N | Q | T | L | V | - | S | F | D | - | - | - | - | - | H | P | L | T | D | G | D | E | V | A | F | F | P | P | V | T | G | G |
| - | M | V | I | G | M | K | F | T | V | I | T | - | - | - | D | D | - | - | - | - | - | - | - | G | K | K | I | L | E | S | G | - | - | A | P | R | R | I | K | D | V | L | G | E | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | I | P | I | - | - | - | - | - | - | E | T | V | V | V | K | K | N | G | Q | I | V | - | I | D | E | - | - | - | - | - | E | E | I | F | D | G | D | I | I | E | V | I | R | V | I | Y | G | G |
| - | - | - | - | - | - | - | - | M | L | Q | L | N | G | - | - | - | - | - | - | - | - | - | - | - | - | K | D | V | K | W | K | - | - | D | T | G | T | I | Q | D | L | L | A | S | Y | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | L | E | N | - | - | - | - | - | - | K | I | V | I | V | E | R | N | K | E | I | I | - | G | K | E | - | R | Y | H | E | V | E | L | C | D | R | D | V | I | E | I | V | H | F | V | G | G | - |
| - | - | - | - | - | - | P | K | V | N | L | Y | A | T | F | R | D | L | T | G | - | - | - | K | S | Q | L | E | L | P | G | A | - | - | - | - | - | T | V | G | E | V | L | E | N | L | V | R | A | Y | - | - | - | P | A | L | K | E | E | L | F | E | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | G | L | A | E | R | V | S | V | F | L | E | G | R | D | - | - | V | R | Y | L | Q | G | L | S | T | P | L | S | P | G | A | T | L | D | L | F | P | P | V | A | G | G |
| - | - | - | - | - | V | K | V | K | V | Y | F | A | R | F | R | Q | L | A | G | - | - | - | V | D | E | E | E | I | E | L | P | - | - | E | G | A | R | V | R | D | L | I | E | E | I | K | K | R | H | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | F | E | E | V | F | G | E | G | Y | D | D | - | - | - | - | - | - | A | D | V | N | I | A | V | N | G | R | Y | V | - | S | W | D | - | - | - | - | - | E | E | L | - | D | G | D | V | V | G | V | F | P | P | V | - | - | - |
| - | - | - | - | - | - | - | - | Q | I | L | F | N | D | - | - | - | - | - | - | - | - | - | - | - | - | Q | A | M | Q | C | A | - | - | A | G | Q | T | V | H | E | L | L | E | Q | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | Q | R | Q | - | - | - | - | - | - | A | G | A | A | L | A | I | N | Q | Q | I | V | - | P | R | E | - | Q | W | A | Q | H | I | V | Q | D | G | D | Q | I | L | L | F | Q | V | I | A | G | G |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V | W | L | N | G | E | P | R | P | L | E | G | K | T | L | K | E | V | L | E | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | V | E | L | K | G | V | A | V | L | L | N | E | E | A | F | L | G | L | E | - | - | V | P | D | R | P | L | R | D | G | D | V | V | E | V | V | A | L | Q | - | - | G |
| - | - | N | Q | I | N | I | E | I | A | Y | A | F | P | - | - | - | - | - | - | - | E | R | Y | Y | L | K | S | F | Q | V | D | E | - | - | G | I | T | V | Q | T | A | I | T | Q | S | G | I | L | S | Q | F | P | E | I | D | L | S | T | N | K | I | G | I | F | S | R | P | I | K | L | T | D | V | L | K | E | G | D | R | I | E | I | Y | R | P | L | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M | S | N | H | N | H | I | T | V | Q | F | A | G | G | C | E | L | L | F | A | - | - | K | Q | T | S | L | Q | L | D | G | V | V | P | T | G | T | N | L | N | G | L | V | Q | L | L | K | T | N | Y | V | K | E | R | - | - | P | D | L | L | V | D | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | G | Q | T | L | R | P | G | I | L | V | L | V | N | S | C | D | - | - | A | E | V | V | G | G | M | D | Y | V | L | N | D | G | D | T | V | E | F | I | S | T | L | H | G | G |
| - | - | - | M | V | N | V | K | V | E | F | L | G | G | L | D | A | I | F | G | - | - | K | Q | R | V | H | K | I | K | M | D | K | E | D | P | V | T | V | G | D | L | I | D | H | I | V | S | T | M | I | N | N | P | N | D | V | S | I | F | I | E | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | S | I | R | P | G | I | I | T | L | I | N | D | T | D | - | - | W | E | L | E | G | E | K | D | Y | I | L | E | D | G | D | I | I | S | F | T | S | T | L | H | G | G |
| - | - | - | K | V | K | I | E | L | K | F | L | G | G | L | E | S | Y | L | E | D | K | S | K | N | Y | V | T | L | E | I | D | S | - | K | E | L | N | F | E | N | L | I | A | F | I | R | D | N | I | I | E | K | K | - | - | - | F | V | F | S | D | Y | D | D | E | K | L | C | K | V | V | D | N | K | E | Y | S | N | Y | N | L | K | D | K | A | K | I | K | P | G | I | I | V | L | V | N | E | Y | D | - | - | W | E | I | L | G | T | Y | S | Y | Q | I | K | N | D | D | - | I | C | F | L | S | T | - | - | - | - |
