solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1lqba- ( 1 ) mdVfLmIrrh-k-------
d1t0ya- ( 1 ) m------------tevYdleittna--td----
d1v2ya1 ( 8 ) mtvrvckm-------dg
d1we6a1 ( 8 ) kfdesa------lvpedqflaqhp-------gpatirvskpnendg----
d1wf9a1 ( 8 ) tmlrvrSrd--------
d1wfya1 ( 8 ) dqevrlenritFqlelvgl--e----
d1wgha1 ( 8 ) msshvpadminlrlilv-------sg
d1wgya1 ( 8 ) e-eifChvyite-------h
d1wxaa1 ( 8 ) sggtlriyAdslk---pni
d1x1ma1 ( 8 ) m---slSdwhlavkladqp----l-------apksilqlpetelge----
d2al6a3 ( 31 ) g-----amervLkVfHyfensseptt
d2c5lc1 (2135 ) esffVqVh--dVS---peq
d2cr5a1 ( 8 ) evpdlpeePsetaeevvtvAlrCpn---g----
d2dhza1 ( 8 ) d-eifcrvYmpd-------h
d2m4na1 ( 2 ) mfggslkvyGGeivpt---r
d3kuza1 (1198 ) tvaLnVvFei--penadv
d4e74a- (1497 ) yktlvLsCvspp--------
d4x57b- ( 4 ) eesidIkFrLydg-------
d6yooa1 ( 1 ) mFqykedhp---feyrkkeGekirkkypdrvPvivekApkArv----
bbbbb
60 70 80 90 100
d1lqba- ( 12 ) tti---ftdakesstVfeLKriVegilk---------------rppdeQr
d1t0ya- ( 16 ) fpm---ekkypagmslndlKkklelvvg---------------ttvdsMr
d1v2ya1 ( 18 ) evm---pvvvvqnAtvldlkkaiqryvqlkqereggvqhiswsyvwrtyh
d1we6a1 ( 41 ) qfm-eitvqsls-envgslKekiageiq---------------ipankqk
d1wf9a1 ( 17 ) gle-rvsvdgp-hitvsqlKtliqdqlq---------------ipihnqt
d1wfya1 ( 28 ) rvv-risa--kptkrlqeAlqpilakhg---------------lsldqvv
d1wgha1 ( 27 ) ktk---eflFspndsAsdiAkhvydnwpmdweeeq-------vsspnilr
d1wgya1 ( 20 ) syv---svkakvssiAqeilkvvAekiq---------------yaeedla
d1wxaa1 ( 24 ) pyk---tillsttDtAdfAvaeSlekYg----lek--------enpkdYc
d1x1ma1 ( 40 ) y--------slggysisflkqliAgklq------------esvpdpelid
d2al6a3 ( 52 ) wAs---iirhgdatdVrgIIqkIVdchkvk--------------nvacyG
d2c5lc1 (2149 ) prt---vIkAprvStAqdVIqqTLckAkySlsIls-------npnpsdYv
d2cr5a1 ( 34 ) rvl---rrrFfkswnSqvlldWMmkv-g---------------yhkslyr
d2dhza1 ( 20 ) syv---tirsrlsasvqdilgsvteklq-------yseepagred--sli
d2m4na1 ( 19 ) pyv---silAeinenAdrilgaAlekYg-------leh------skddFi
d3kuza1 (1217 ) cni---sVnVldcDtIgqAkekIfqaFlskn---gspY-gL---qlneIg
d4e74a- (1514 ) -ev---pvkIlncdtItqVK-kIldaIfk-----nvpcshr---paadmd
d4x57b- ( 17 ) sdi--GpfrysaastVdfLKqrVvsdWp---------gktvvPkgineVk
d6yooa1 ( 42 ) pdldkrkylvpsdltvgqFyflirkrih---------lrp-----edalf
b bb aaaaaaaaaaa b
110 120 130 140 150
d1lqba- ( 44 ) Lykdd------------------qlLddgk-tLgecgFts-qtArpqapa
d1t0ya- ( 48 ) iqlfd-gdd----------qlkgeltdGak-slkdlgvrd--------gy
d1v2ya1 ( 65 ) ltsa------geklted-----------rk-klrdygir--------nrd
d1we6a1 ( 74 ) lsGk-a----------------gflkd-nm-slAhynvGa--------ge
d1wf9a1 ( 50 ) lStn-rnlllAk--spsdflaftdMadpnl-risslnlah--------gs
d1wfya1 ( 60 ) lhrp-gek--------------qpmdl-en-pvssva-----------sq
d1wgha1 ( 67 ) liyq------------------grflhgnv-tlgal--klp----fgktt
d1wgya1 ( 52 ) lvAit-f------------sgekhelqpnd-lvisksl--------easg
d1wxaa1 ( 59 ) iArvm-lppgaqh--sdergAkeiilddde-cPlqifrewpsdkg--i-l
d1x1ma1 ( 70 ) liYc-g----------------rklkd-dq-tldfygiqp--------gs
d2al6a3 ( 85 ) LrLsH-lqse-----------evhwLhldm-gVsnVrekfelahppee-W
d2c5lc1 (2189 ) Lleev-v------------kssqrvLldqe-cVfqAqskwk---G--a-G
d2cr5a1 ( 65 ) lstsf--------------prraleveggs-sledigitv--------dt
d2dhza1 ( 58 ) lvAvs-s------------sgekvllqptedcvFtalg---------ins
d2m4na1 ( 53 ) lvevs-ndddrksms-dlreidgrpippte-cPlfeMtarsgnGengfds
d3kuza1 (1258 ) Lelmg------keLldidssSvilgitklN-tIghY-Is--------ngs
d4e74a- (1553 ) Lewrq-gsgarmiLqdedittk------lN-tLahyqVp--------dgs
d4x57b- ( 57 ) Lissg------------------kiLennk-tvgqck-tp-fgdiaggvi
d6yooa1 ( 78 ) FFvnn------------------tiPptsa-tMgqlYe---dnheeDyFl
bb
160 170 180
d1lqba- ( 74 ) tVGLAfraddtfealciepfssppelpdvmk
d1t0ya- ( 78 ) rihAvDvtggned
d1v2ya1 ( 89 ) evsfikkl--------------------gqk
d1we6a1 ( 97 ) iltlslre--------r
d1wf9a1 ( 88 ) mvylAYegerti---rg
d1wfya1 ( 82 ) tlvldtppdakmsears
d1wgha1 ( 92 ) vMhlvAre--t----------lpepnsqgqr
d1wgya1 ( 80 ) riyvYrkd--ladtlnpfae----------n
d1wxaa1 ( 102 ) vFqlkrrp--p
d1x1ma1 ( 93 ) tvhvlrks--------w
d2al6a3 ( 121 ) kyELriry--l----------p
d2c5lc1 (2230 ) kFILklke--q
d2cr5a1 ( 92 ) vlnveeke-----qss--------------q
d2dhza1 ( 86 ) hlFACtrd--syealvplpeeiqvspgdtei
d2m4na1 ( 100 ) FlAikrkp--h
d3kuza1 (1299 ) tIkVfk
d4e74a- (1594 ) vVaLvskq----------------------v
d4x57b- ( 86 ) vMhVvVqps
d6yooa1 ( 106 ) YvAYSdesvygk
bbbbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Elongin B | Ubiquitin-related | Human (Homo sapiens) [TaxId: 9606] | 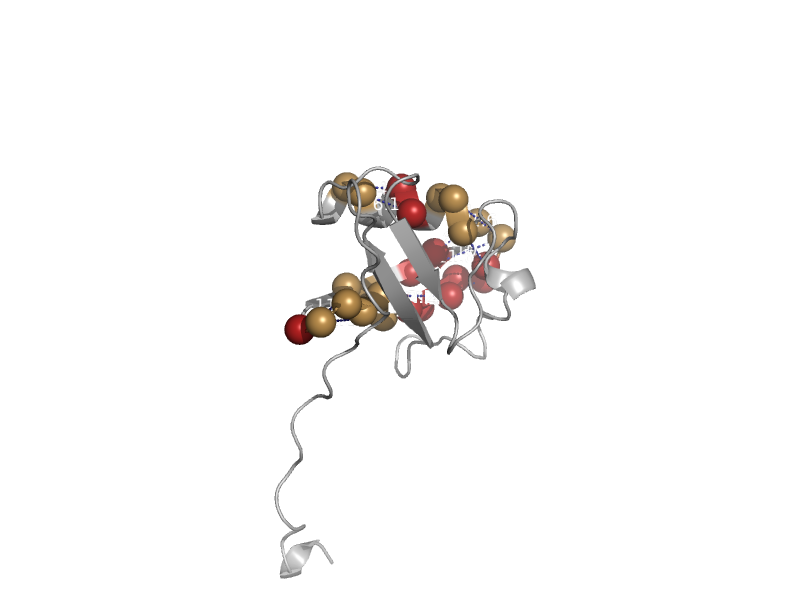 | view | |
| Ubiquitin-like domain of tubulin folding cofactor B | Ubiquitin-related | Nematode (Caenorhabditis elegans) [TaxId: 6239] | 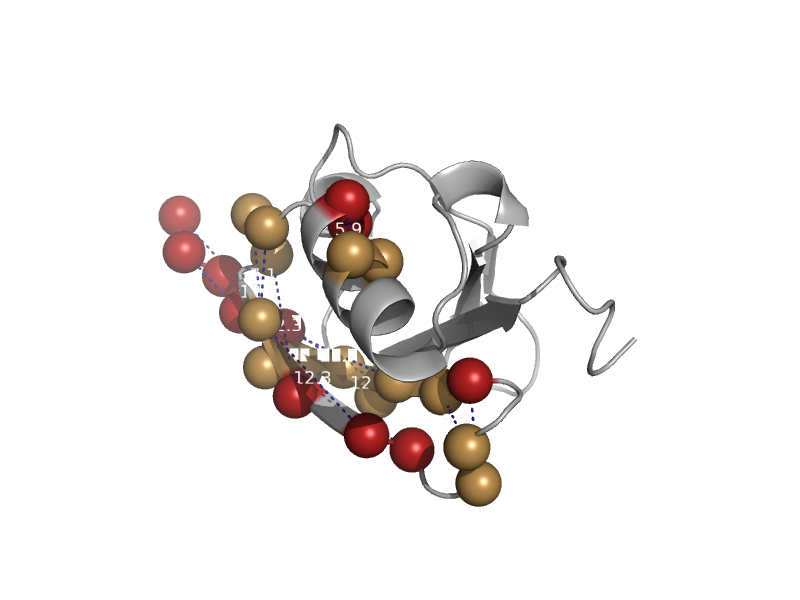 | view | |
| Ubiquitin-like protein 3300001g02rik | Ubiquitin-related | Mouse (Mus musculus) [TaxId: 10090] | 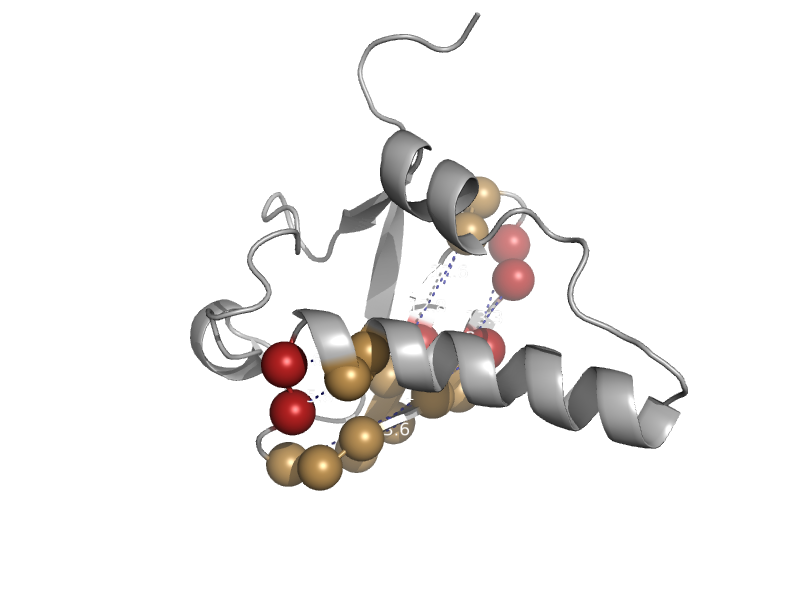 | view | |
| Splicing factor 3 subunit 1, C-terminal domain | Ubiquitin-related | Thale cress (Arabidopsis thaliana) [TaxId: 3702] | 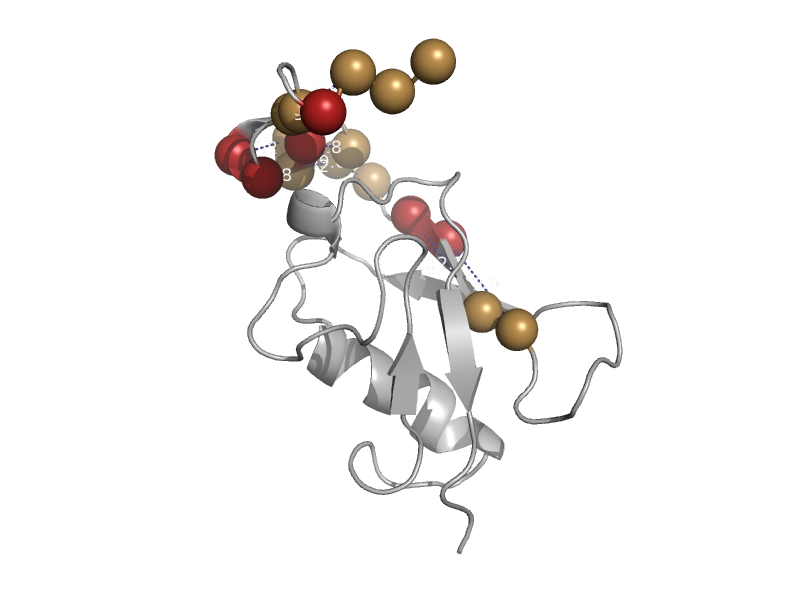 | view | |
| NPL4-like protein 1 | Ubiquitin-related | Thale cress (Arabidopsis thaliana) [TaxId: 3702] | 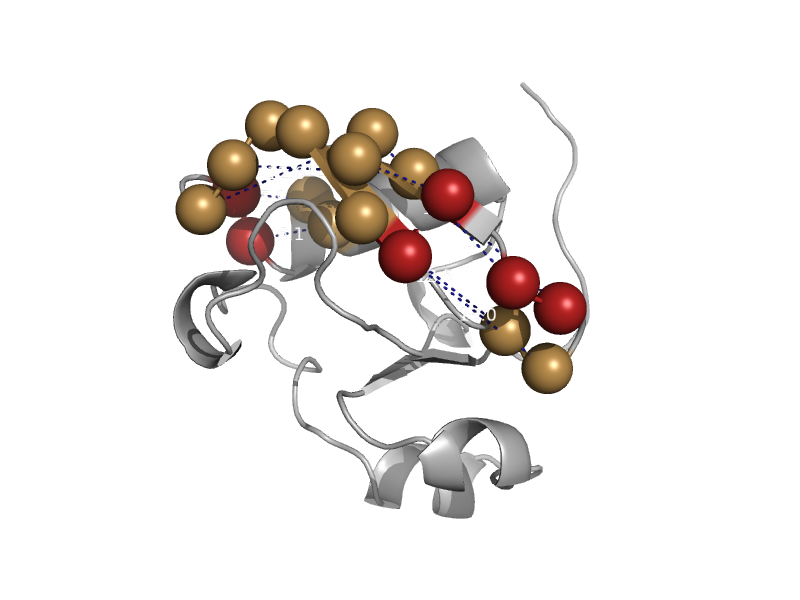 | view | |
| Regulator of G-protein signaling 14, RGS14 | Ras-binding domain, RBD | Mouse (Mus musculus) [TaxId: 10090] | 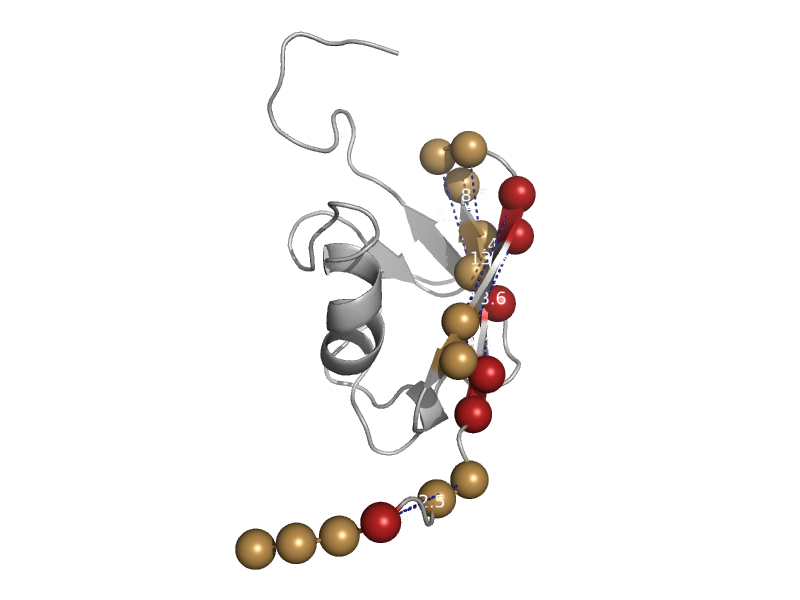 | view | |
| Ubiquitin-like protein 3, Ubl3 | Ubiquitin-related | Mouse (Mus musculus) [TaxId: 10090] | 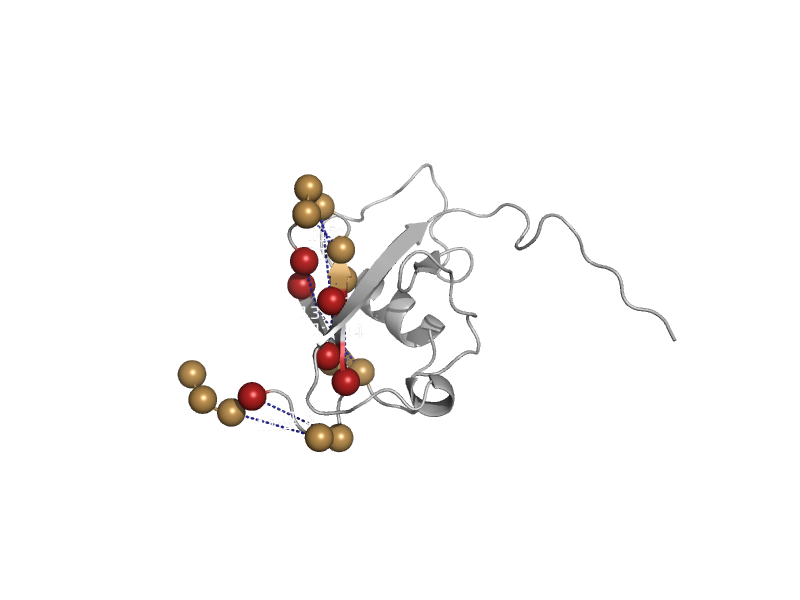 | view | |
| Rap guanine nucleotide exchange factor 5, RapGEF5 | Ras-binding domain, RBD | Human (Homo sapiens) [TaxId: 9606] | 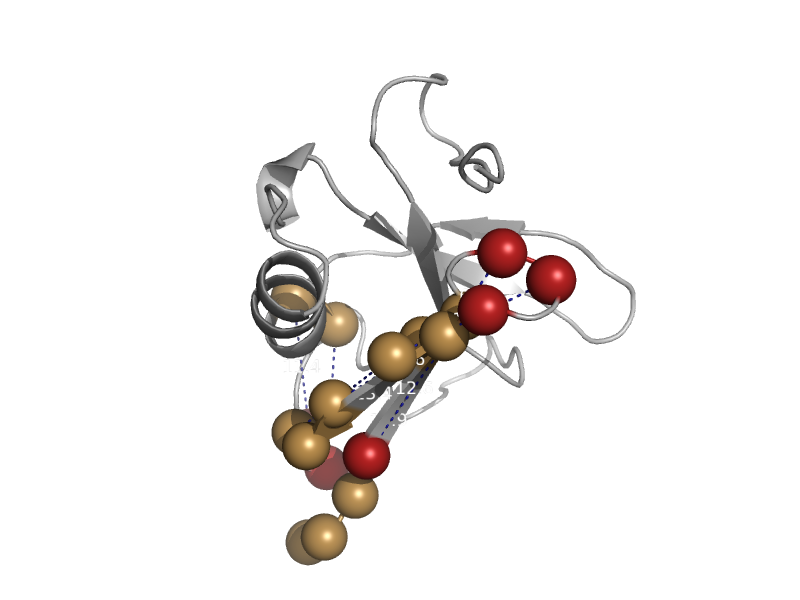 | view | |
| Afadin | Ras-binding domain, RBD | Mouse (Mus musculus) [TaxId: 10090] |  | view | |
| Ubiquitin-like protein 7 | Ubiquitin-related | Mouse (Mus musculus) [TaxId: 10090] |  | view | |
| Focal adhesion kinase 1 | First domain of FERM | Chicken (Gallus gallus) [TaxId: 9031] |  | view | |
| Phospholipase C-epsilon-1 | Ras-binding domain, RBD | Human (Homo sapiens) [TaxId: 9606] |  | view | |
| UBX domain-containing protein 6 (Reproduction 8) | UBX domain | Mouse (Mus musculus) [TaxId: 10090] |  | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 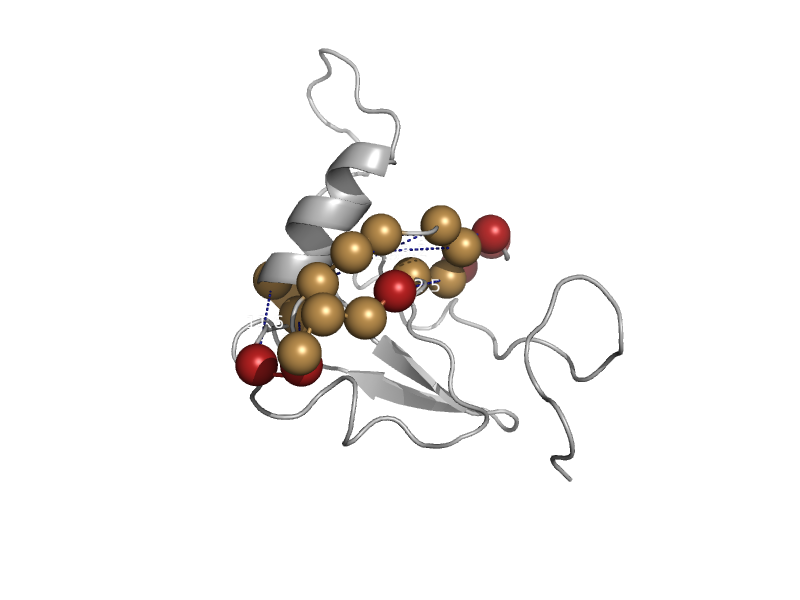 | view | |
| automated matches | automated matches | Nematode (Caenorhabditis elegans) [TaxId: 6239] | 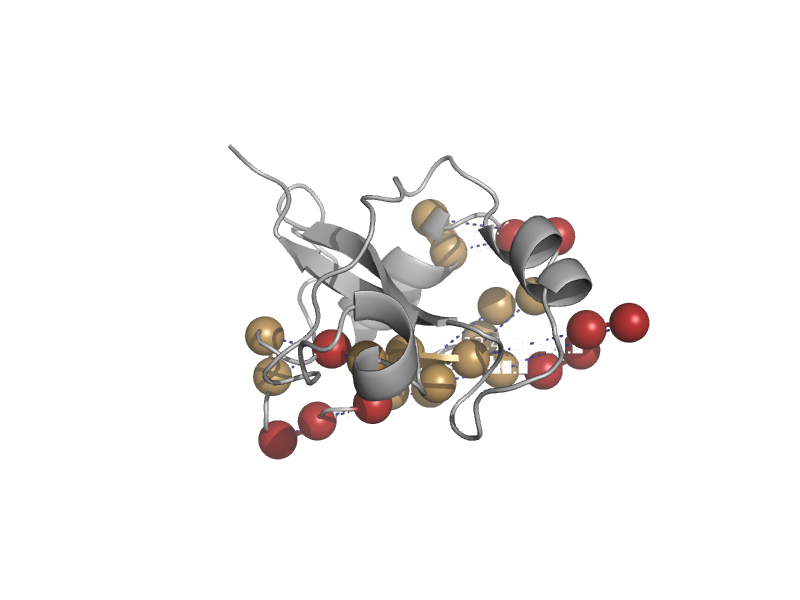 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] |  | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] |  | view | |
| automated matches | automated matches | Thale cress (Arabidopsis thaliana) [TaxId: 3702] | 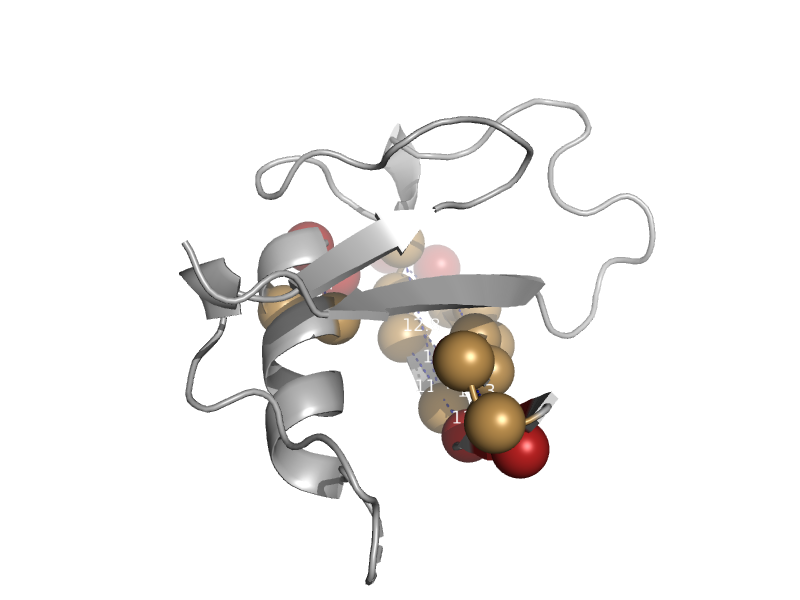 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 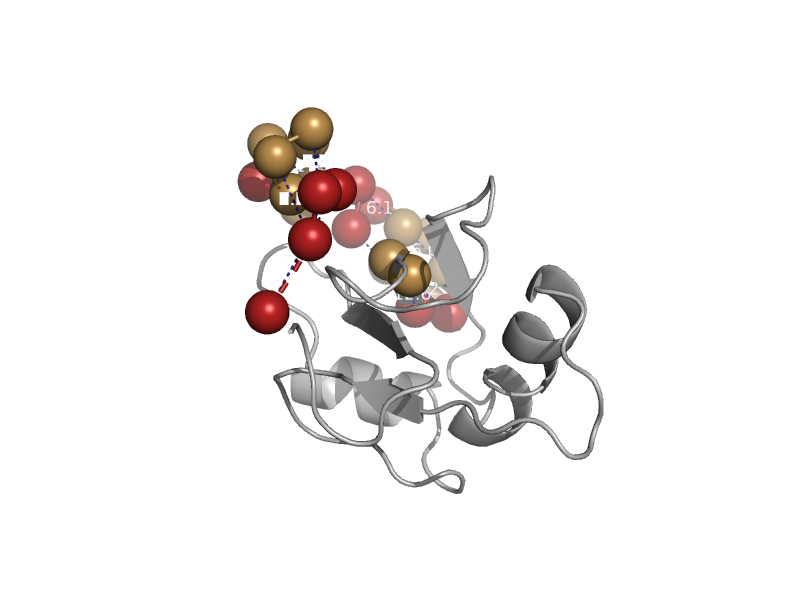 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | D | V | F | L | M | I | R | R | H | - | K | - | - | - | - | - | - | - | T | T | I | - | - | - | F | T | D | A | K | E | S | S | T | V | F | E | L | K | R | I | V | E | G | I | L | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | P | P | D | E | Q | R | L | Y | K | D | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | L | L | D | D | G | K | - | T | L | G | E | C | G | F | T | S | - | Q | T | A | R | P | Q | A | P | A | T | V | G | L | A | F | R | A | D | D | T | F | E | A | L | C | I | E | P | F | S | S | P | P | E | L | P | D | V | M | K |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | - | - | - | - | - | - | - | - | - | - | - | - | T | E | V | Y | D | L | E | I | T | T | N | A | - | - | T | D | - | - | - | - | F | P | M | - | - | - | E | K | K | Y | P | A | G | M | S | L | N | D | L | K | K | K | L | E | L | V | V | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | T | V | D | S | M | R | I | Q | L | F | D | - | G | D | D | - | - | - | - | - | - | - | - | - | - | Q | L | K | G | E | L | T | D | G | A | K | - | S | L | K | D | L | G | V | R | D | - | - | - | - | - | - | - | - | G | Y | R | I | H | A | V | D | V | T | G | G | N | E | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | T | V | R | V | C | K | M | - | - | - | - | - | - | - | D | G | E | V | M | - | - | - | P | V | V | V | V | Q | N | A | T | V | L | D | L | K | K | A | I | Q | R | Y | V | Q | L | K | Q | E | R | E | G | G | V | Q | H | I | S | W | S | Y | V | W | R | T | Y | H | L | T | S | A | - | - | - | - | - | - | G | E | K | L | T | E | D | - | - | - | - | - | - | - | - | - | - | - | R | K | - | K | L | R | D | Y | G | I | R | - | - | - | - | - | - | - | - | N | R | D | E | V | S | F | I | K | K | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | Q | K |
| K | F | D | E | S | A | - | - | - | - | - | - | L | V | P | E | D | Q | F | L | A | Q | H | P | - | - | - | - | - | - | - | G | P | A | T | I | R | V | S | K | P | N | E | N | D | G | - | - | - | - | Q | F | M | - | E | I | T | V | Q | S | L | S | - | E | N | V | G | S | L | K | E | K | I | A | G | E | I | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | I | P | A | N | K | Q | K | L | S | G | K | - | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | F | L | K | D | - | N | M | - | S | L | A | H | Y | N | V | G | A | - | - | - | - | - | - | - | - | G | E | I | L | T | L | S | L | R | E | - | - | - | - | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | M | L | R | V | R | S | R | D | - | - | - | - | - | - | - | - | G | L | E | - | R | V | S | V | D | G | P | - | H | I | T | V | S | Q | L | K | T | L | I | Q | D | Q | L | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | I | P | I | H | N | Q | T | L | S | T | N | - | R | N | L | L | L | A | K | - | - | S | P | S | D | F | L | A | F | T | D | M | A | D | P | N | L | - | R | I | S | S | L | N | L | A | H | - | - | - | - | - | - | - | - | G | S | M | V | Y | L | A | Y | E | G | E | R | T | I | - | - | - | R | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | Q | E | V | R | L | E | N | R | I | T | F | Q | L | E | L | V | G | L | - | - | E | - | - | - | - | R | V | V | - | R | I | S | A | - | - | K | P | T | K | R | L | Q | E | A | L | Q | P | I | L | A | K | H | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | S | L | D | Q | V | V | L | H | R | P | - | G | E | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | P | M | D | L | - | E | N | - | P | V | S | S | V | A | - | - | - | - | - | - | - | - | - | - | - | S | Q | T | L | V | L | D | T | P | P | D | A | K | M | S | E | A | R | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | S | S | H | V | P | A | D | M | I | N | L | R | L | I | L | V | - | - | - | - | - | - | - | S | G | K | T | K | - | - | - | E | F | L | F | S | P | N | D | S | A | S | D | I | A | K | H | V | Y | D | N | W | P | M | D | W | E | E | E | Q | - | - | - | - | - | - | - | V | S | S | P | N | I | L | R | L | I | Y | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | R | F | L | H | G | N | V | - | T | L | G | A | L | - | - | K | L | P | - | - | - | - | F | G | K | T | T | V | M | H | L | V | A | R | E | - | - | T | - | - | - | - | - | - | - | - | - | - | L | P | E | P | N | S | Q | G | Q | R |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | - | E | I | F | C | H | V | Y | I | T | E | - | - | - | - | - | - | - | H | S | Y | V | - | - | - | S | V | K | A | K | V | S | S | I | A | Q | E | I | L | K | V | V | A | E | K | I | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | A | E | E | D | L | A | L | V | A | I | T | - | F | - | - | - | - | - | - | - | - | - | - | - | - | S | G | E | K | H | E | L | Q | P | N | D | - | L | V | I | S | K | S | L | - | - | - | - | - | - | - | - | E | A | S | G | R | I | Y | V | Y | R | K | D | - | - | L | A | D | T | L | N | P | F | A | E | - | - | - | - | - | - | - | - | - | - | N |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | G | G | T | L | R | I | Y | A | D | S | L | K | - | - | - | P | N | I | P | Y | K | - | - | - | T | I | L | L | S | T | T | D | T | A | D | F | A | V | A | E | S | L | E | K | Y | G | - | - | - | - | L | E | K | - | - | - | - | - | - | - | - | E | N | P | K | D | Y | C | I | A | R | V | M | - | L | P | P | G | A | Q | H | - | - | S | D | E | R | G | A | K | E | I | I | L | D | D | D | E | - | C | P | L | Q | I | F | R | E | W | P | S | D | K | G | - | - | I | - | L | V | F | Q | L | K | R | R | P | - | - | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M | - | - | - | S | L | S | D | W | H | L | A | V | K | L | A | D | Q | P | - | - | - | - | L | - | - | - | - | - | - | - | A | P | K | S | I | L | Q | L | P | E | T | E | L | G | E | - | - | - | - | Y | - | - | - | - | - | - | - | - | S | L | G | G | Y | S | I | S | F | L | K | Q | L | I | A | G | K | L | Q | - | - | - | - | - | - | - | - | - | - | - | - | E | S | V | P | D | P | E | L | I | D | L | I | Y | C | - | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | K | L | K | D | - | D | Q | - | T | L | D | F | Y | G | I | Q | P | - | - | - | - | - | - | - | - | G | S | T | V | H | V | L | R | K | S | - | - | - | - | - | - | - | - | W | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | - | - | - | - | - | A | M | E | R | V | L | K | V | F | H | Y | F | E | N | S | S | E | P | T | T | W | A | S | - | - | - | I | I | R | H | G | D | A | T | D | V | R | G | I | I | Q | K | I | V | D | C | H | K | V | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | V | A | C | Y | G | L | R | L | S | H | - | L | Q | S | E | - | - | - | - | - | - | - | - | - | - | - | E | V | H | W | L | H | L | D | M | - | G | V | S | N | V | R | E | K | F | E | L | A | H | P | P | E | E | - | W | K | Y | E | L | R | I | R | Y | - | - | L | - | - | - | - | - | - | - | - | - | - | P | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | S | F | F | V | Q | V | H | - | - | D | V | S | - | - | - | P | E | Q | P | R | T | - | - | - | V | I | K | A | P | R | V | S | T | A | Q | D | V | I | Q | Q | T | L | C | K | A | K | Y | S | L | S | I | L | S | - | - | - | - | - | - | - | N | P | N | P | S | D | Y | V | L | L | E | E | V | - | V | - | - | - | - | - | - | - | - | - | - | - | - | K | S | S | Q | R | V | L | L | D | Q | E | - | C | V | F | Q | A | Q | S | K | W | K | - | - | - | G | - | - | A | - | G | K | F | I | L | K | L | K | E | - | - | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | V | P | D | L | P | E | E | P | S | E | T | A | E | E | V | V | T | V | A | L | R | C | P | N | - | - | - | G | - | - | - | - | R | V | L | - | - | - | R | R | R | F | F | K | S | W | N | S | Q | V | L | L | D | W | M | M | K | V | - | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | H | K | S | L | Y | R | L | S | T | S | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | R | R | A | L | E | V | E | G | G | S | - | S | L | E | D | I | G | I | T | V | - | - | - | - | - | - | - | - | D | T | V | L | N | V | E | E | K | E | - | - | - | - | - | Q | S | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | - | E | I | F | C | R | V | Y | M | P | D | - | - | - | - | - | - | - | H | S | Y | V | - | - | - | T | I | R | S | R | L | S | A | S | V | Q | D | I | L | G | S | V | T | E | K | L | Q | - | - | - | - | - | - | - | Y | S | E | E | P | A | G | R | E | D | - | - | S | L | I | L | V | A | V | S | - | S | - | - | - | - | - | - | - | - | - | - | - | - | S | G | E | K | V | L | L | Q | P | T | E | D | C | V | F | T | A | L | G | - | - | - | - | - | - | - | - | - | I | N | S | H | L | F | A | C | T | R | D | - | - | S | Y | E | A | L | V | P | L | P | E | E | I | Q | V | S | P | G | D | T | E | I |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | F | G | G | S | L | K | V | Y | G | G | E | I | V | P | T | - | - | - | R | P | Y | V | - | - | - | S | I | L | A | E | I | N | E | N | A | D | R | I | L | G | A | A | L | E | K | Y | G | - | - | - | - | - | - | - | L | E | H | - | - | - | - | - | - | S | K | D | D | F | I | L | V | E | V | S | - | N | D | D | D | R | K | S | M | S | - | D | L | R | E | I | D | G | R | P | I | P | P | T | E | - | C | P | L | F | E | M | T | A | R | S | G | N | G | E | N | G | F | D | S | F | L | A | I | K | R | K | P | - | - | H | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | V | A | L | N | V | V | F | E | I | - | - | P | E | N | A | D | V | C | N | I | - | - | - | S | V | N | V | L | D | C | D | T | I | G | Q | A | K | E | K | I | F | Q | A | F | L | S | K | N | - | - | - | G | S | P | Y | - | G | L | - | - | - | Q | L | N | E | I | G | L | E | L | M | G | - | - | - | - | - | - | K | E | L | L | D | I | D | S | S | S | V | I | L | G | I | T | K | L | N | - | T | I | G | H | Y | - | I | S | - | - | - | - | - | - | - | - | N | G | S | T | I | K | V | F | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | K | T | L | V | L | S | C | V | S | P | P | - | - | - | - | - | - | - | - | - | E | V | - | - | - | P | V | K | I | L | N | C | D | T | I | T | Q | V | K | - | K | I | L | D | A | I | F | K | - | - | - | - | - | N | V | P | C | S | H | R | - | - | - | P | A | A | D | M | D | L | E | W | R | Q | - | G | S | G | A | R | M | I | L | Q | D | E | D | I | T | T | K | - | - | - | - | - | - | L | N | - | T | L | A | H | Y | Q | V | P | - | - | - | - | - | - | - | - | D | G | S | V | V | A | L | V | S | K | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | E | S | I | D | I | K | F | R | L | Y | D | G | - | - | - | - | - | - | - | S | D | I | - | - | G | P | F | R | Y | S | A | A | S | T | V | D | F | L | K | Q | R | V | V | S | D | W | P | - | - | - | - | - | - | - | - | - | G | K | T | V | V | P | K | G | I | N | E | V | K | L | I | S | S | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | I | L | E | N | N | K | - | T | V | G | Q | C | K | - | T | P | - | F | G | D | I | A | G | G | V | I | V | M | H | V | V | V | Q | P | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | M | F | Q | Y | K | E | D | H | P | - | - | - | F | E | Y | R | K | K | E | G | E | K | I | R | K | K | Y | P | D | R | V | P | V | I | V | E | K | A | P | K | A | R | V | - | - | - | - | P | D | L | D | K | R | K | Y | L | V | P | S | D | L | T | V | G | Q | F | Y | F | L | I | R | K | R | I | H | - | - | - | - | - | - | - | - | - | L | R | P | - | - | - | - | - | E | D | A | L | F | F | F | V | N | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | I | P | P | T | S | A | - | T | M | G | Q | L | Y | E | - | - | - | D | N | H | E | E | D | Y | F | L | Y | V | A | Y | S | D | E | S | V | Y | G | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
