solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1qgoa- ( 2 ) kkALLVVSfGTsy----------------------------------
d2h1va- ( 2 ) srkkMGLLVMaygTPykeedIerYythirr-------------grkPepe
d2xvxa- ( 8 ) qktGILLVAfGTsv----------------------------------
d2xwsa1 ( 1 )
d3hcna- ( 65 ) rkpkTGILMLNmGGPetlgdVhdFLlrlfldrDlmtlpiQnklApfiAkr
bbbbbbb
60 70 80 90 100
d1qgoa- ( 15 ) ------------------hdtcekNIvaCErdLaasCp-dRdlfrAFtsg
d2h1va- ( 39 ) mlqdlkdrYeaiggIspLAqiteqQAhnLeqhLneiqde-ItFkAYiGLA
d2xvxa- ( 22 ) ------------------eeArpa-ldkMGdrVraahp-dipVrwAytak
d2xwsa1 ( 1 )
d3hcna- ( 115 ) rtpkiqeqyrriggGspIkiwTskqGegMvklLdelSpntapHkyYiGFr
aa aaaaaaaaaa bbbbb
110 120 130 140 150
d1qgoa- ( 46 ) miirkLrqrdgididtPlqALqkLaaqgYqdVAIQSLhiingde-yekIv
d2h1va- ( 88 ) hIe-----------pfIedAVaeMhkdgIteAVSIVLaPHfStfsVqsYn
d2xvxa- ( 52 ) mirakLrae-giaApspaeaLagMaeegFthVAVQSLhtipgee-FhgLl
d2xwsa1 ( 1 )
d3hcna- ( 165 ) Yvh-----------plTeeAIeeMerdgLerAIAFTQyPQyScstTgSSL
aaaaaaaaaa bbbbbb aaaaa
160 170 180 190 200
d1qgoa- ( 95 ) reVqllrp-l---Ft-rLtlGvPLLsshndyvqLMqALrqqMp-slr---
d2h1va- ( 127 ) krAkeeAeklg---gLtItSVesWydepkFvtYWvdrVkety-asMpede
d2xvxa- ( 100 ) etAhafqg-lpkgLt-rVSVGlPLIgttadaeaVAeaLvaslpadrk---
d2xwsa1 ( 1 )
d3hcna- ( 204 ) NAIyryYnqvgrkptMkWsTIDrWpthhlLiqCFAdHIlkel-dhFplek
aaaaaa bbbb aaaaaaaaaaaa
210 220 230 240 250
d1qgoa- ( 136 ) -qtekVVFMGhgas-------hhAfaaYacLdhmMtaq---rfpArVGAV
d2h1va- ( 173 ) renAMLIVSAhSlpekIkefgDpypdqLheSAklIAegAgVs-eyavGWQ
d2xvxa- ( 145 ) -pgePVVFMGhGtp-------hpadicYpgLqyyLwrl---dpdLLVGTv
d2xwsa1 ( 1 ) mrrGLVIVGhgsql------nhyrevMelhrkriees-gafdeVkiAFA
d3hcna- ( 253 ) rseVVILFSAhSlpmsvvnrgDpypqeVsaTVqkVMerLeycNpyrlVWq
bbbbbb aaaaaaaaaaaaaaa bbbbb
260 270 280 290 300
d1qgoa- ( 175 ) e--------sype-VdiLidsLrdegVtgVhLMPL-mlvagdhAindMas
d2h1va- ( 222 ) segntpdpwLgpdVqdlTrdlfeqkgYqAFVYVPVGfVAdhleVlydNdy
d2xvxa- ( 184 ) e--------gsps-fdnVmaeLdvrkakrVwLmPL-mavAgdhArndMAg
d2xwsa1 ( 43 ) Ar------krrpmPdeAIreM----nCdiIyVvPl-fisyg---------
d3hcna- ( 303 ) sk-vgpmpwlgpqTdesIkglCer-grkNILLVPIAftsdhieTlYeldi
aaaaaaaaa bbbbb a
310 320 330 340 350
d1qgoa- ( 215 ) ddgdSWkmrFna-ag-------------IpAtpwlsGLGenpaIraMFva
d2h1va- ( 272 ) ----eCkvvTdd-ig-------------A-syyrpeMPnakpeFIdALAt
d2xvxa- ( 224 ) deddSWtsqLar-rg-------------ieAkpvlhGtAesdaVAaIWlr
d2xwsa1 ( 73 ) ---lhVtedLPdlLgFprgrgikegefegkkVvIcePiGedyfvtyailn
d3hcna- ( 351 ) ----eysqvLakecg-------------venirrAeSLngnplFskALAd
aaaaaaa bbb aaaaaaaaa
360 370 380 390
d1qgoa- ( 251 ) HLhqalnm
d2h1va- ( 303 ) vVlkklgr
d2xvxa- ( 260 ) HLddAlarLn
d2xwsa1 ( 120 ) svfri----------------------------------g
d3hcna- ( 384 ) lVhsHIqsnelCskqltlscplcvnpvcreTksfFtsqql
aaaaa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Cobalt chelatase CbiK | Cobalt chelatase CbiK | Salmonella typhimurium [TaxId: 90371] | 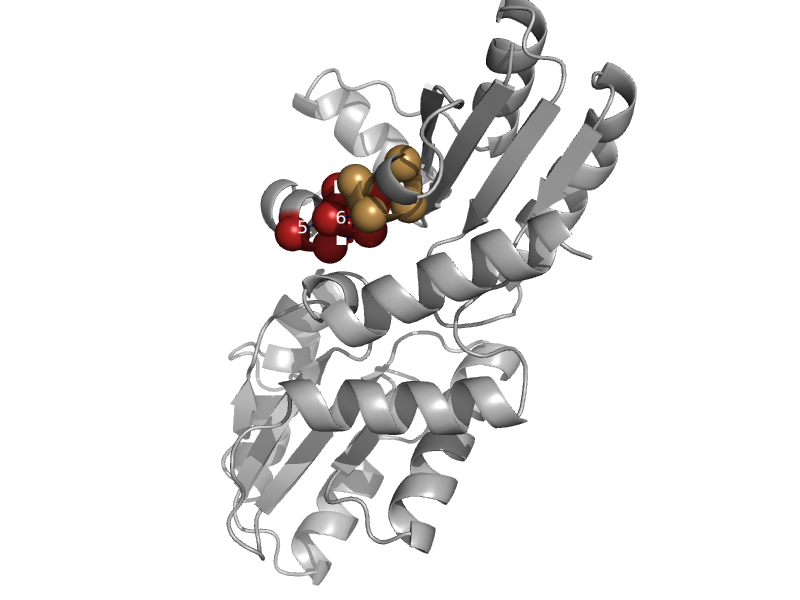 | view | |
| Ferrochelatase | Ferrochelatase | Bacillus subtilis [TaxId: 1423] | 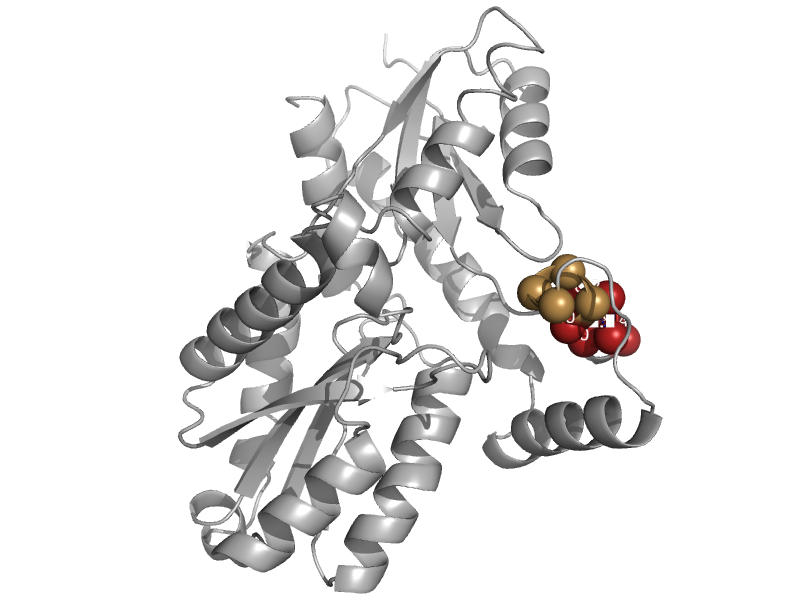 | view | |
| automated matches | automated matches | Desulfovibrio vulgaris [TaxId: 882] | 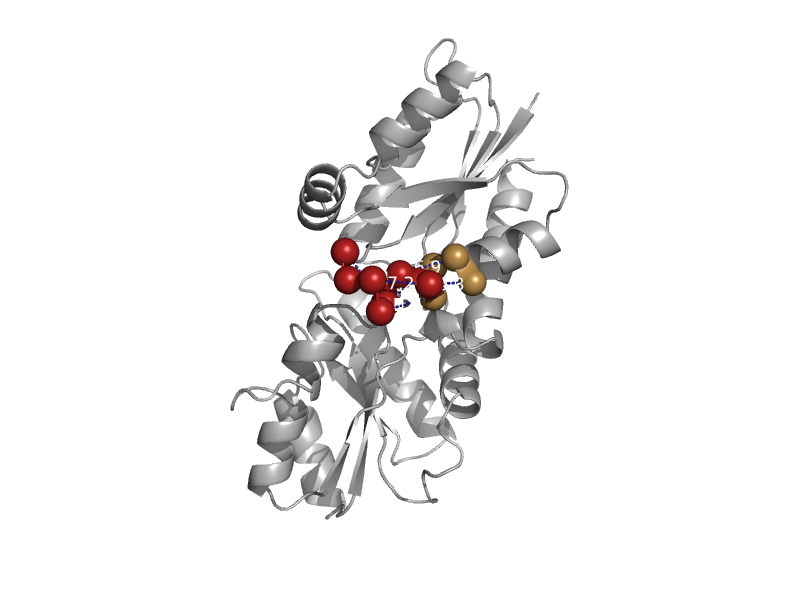 | view | |
| automated matches | CbiX-like | Archaeoglobus fulgidus [TaxId: 2234] | 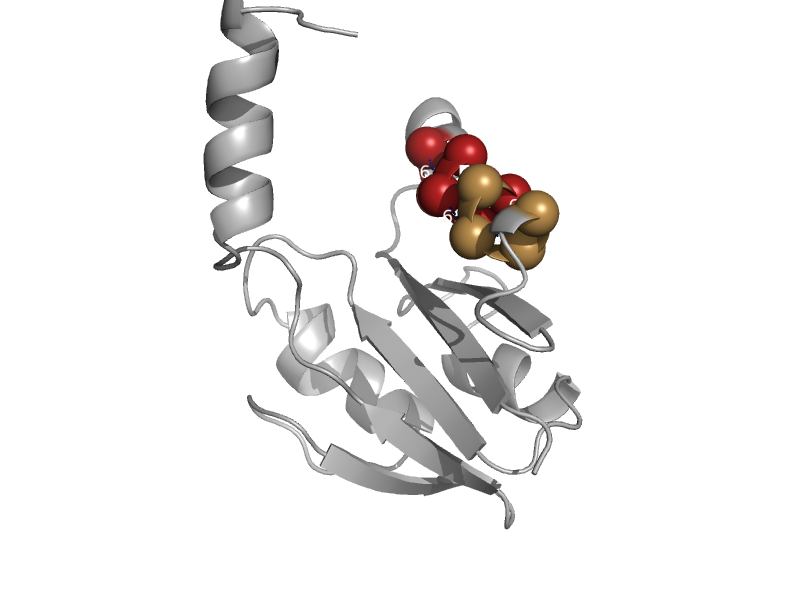 | view | |
| Ferrochelatase | Ferrochelatase | Human (Homo sapiens) [TaxId: 9606] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | 330 | 340 | 350 | 360 | 370 | 380 | 390 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | K | K | A | L | L | V | V | S | F | G | T | S | Y | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | H | D | T | C | E | K | N | I | V | A | C | E | R | D | L | A | A | S | C | P | - | D | R | D | L | F | R | A | F | T | S | G | M | I | I | R | K | L | R | Q | R | D | G | I | D | I | D | T | P | L | Q | A | L | Q | K | L | A | A | Q | G | Y | Q | D | V | A | I | Q | S | L | H | I | I | N | G | D | E | - | Y | E | K | I | V | R | E | V | Q | L | L | R | P | - | L | - | - | - | F | T | - | R | L | T | L | G | V | P | L | L | S | S | H | N | D | Y | V | Q | L | M | Q | A | L | R | Q | Q | M | P | - | S | L | R | - | - | - | - | Q | T | E | K | V | V | F | M | G | H | G | A | S | - | - | - | - | - | - | - | H | H | A | F | A | A | Y | A | C | L | D | H | M | M | T | A | Q | - | - | - | R | F | P | A | R | V | G | A | V | E | - | - | - | - | - | - | - | - | S | Y | P | E | - | V | D | I | L | I | D | S | L | R | D | E | G | V | T | G | V | H | L | M | P | L | - | M | L | V | A | G | D | H | A | I | N | D | M | A | S | D | D | G | D | S | W | K | M | R | F | N | A | - | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | I | P | A | T | P | W | L | S | G | L | G | E | N | P | A | I | R | A | M | F | V | A | H | L | H | Q | A | L | N | M | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| S | R | K | K | M | G | L | L | V | M | A | Y | G | T | P | Y | K | E | E | D | I | E | R | Y | Y | T | H | I | R | R | - | - | - | - | - | - | - | - | - | - | - | - | - | G | R | K | P | E | P | E | M | L | Q | D | L | K | D | R | Y | E | A | I | G | G | I | S | P | L | A | Q | I | T | E | Q | Q | A | H | N | L | E | Q | H | L | N | E | I | Q | D | E | - | I | T | F | K | A | Y | I | G | L | A | H | I | E | - | - | - | - | - | - | - | - | - | - | - | P | F | I | E | D | A | V | A | E | M | H | K | D | G | I | T | E | A | V | S | I | V | L | A | P | H | F | S | T | F | S | V | Q | S | Y | N | K | R | A | K | E | E | A | E | K | L | G | - | - | - | G | L | T | I | T | S | V | E | S | W | Y | D | E | P | K | F | V | T | Y | W | V | D | R | V | K | E | T | Y | - | A | S | M | P | E | D | E | R | E | N | A | M | L | I | V | S | A | H | S | L | P | E | K | I | K | E | F | G | D | P | Y | P | D | Q | L | H | E | S | A | K | L | I | A | E | G | A | G | V | S | - | E | Y | A | V | G | W | Q | S | E | G | N | T | P | D | P | W | L | G | P | D | V | Q | D | L | T | R | D | L | F | E | Q | K | G | Y | Q | A | F | V | Y | V | P | V | G | F | V | A | D | H | L | E | V | L | Y | D | N | D | Y | - | - | - | - | E | C | K | V | V | T | D | D | - | I | G | - | - | - | - | - | - | - | - | - | - | - | - | - | A | - | S | Y | Y | R | P | E | M | P | N | A | K | P | E | F | I | D | A | L | A | T | V | V | L | K | K | L | G | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | Q | K | T | G | I | L | L | V | A | F | G | T | S | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | E | A | R | P | A | - | L | D | K | M | G | D | R | V | R | A | A | H | P | - | D | I | P | V | R | W | A | Y | T | A | K | M | I | R | A | K | L | R | A | E | - | G | I | A | A | P | S | P | A | E | A | L | A | G | M | A | E | E | G | F | T | H | V | A | V | Q | S | L | H | T | I | P | G | E | E | - | F | H | G | L | L | E | T | A | H | A | F | Q | G | - | L | P | K | G | L | T | - | R | V | S | V | G | L | P | L | I | G | T | T | A | D | A | E | A | V | A | E | A | L | V | A | S | L | P | A | D | R | K | - | - | - | - | P | G | E | P | V | V | F | M | G | H | G | T | P | - | - | - | - | - | - | - | H | P | A | D | I | C | Y | P | G | L | Q | Y | Y | L | W | R | L | - | - | - | D | P | D | L | L | V | G | T | V | E | - | - | - | - | - | - | - | - | G | S | P | S | - | F | D | N | V | M | A | E | L | D | V | R | K | A | K | R | V | W | L | M | P | L | - | M | A | V | A | G | D | H | A | R | N | D | M | A | G | D | E | D | D | S | W | T | S | Q | L | A | R | - | R | G | - | - | - | - | - | - | - | - | - | - | - | - | - | I | E | A | K | P | V | L | H | G | T | A | E | S | D | A | V | A | A | I | W | L | R | H | L | D | D | A | L | A | R | L | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | R | R | G | L | V | I | V | G | H | G | S | Q | L | - | - | - | - | - | - | N | H | Y | R | E | V | M | E | L | H | R | K | R | I | E | E | S | - | G | A | F | D | E | V | K | I | A | F | A | A | R | - | - | - | - | - | - | K | R | R | P | M | P | D | E | A | I | R | E | M | - | - | - | - | N | C | D | I | I | Y | V | V | P | L | - | F | I | S | Y | G | - | - | - | - | - | - | - | - | - | - | - | - | L | H | V | T | E | D | L | P | D | L | L | G | F | P | R | G | R | G | I | K | E | G | E | F | E | G | K | K | V | V | I | C | E | P | I | G | E | D | Y | F | V | T | Y | A | I | L | N | S | V | F | R | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G |
| R | K | P | K | T | G | I | L | M | L | N | M | G | G | P | E | T | L | G | D | V | H | D | F | L | L | R | L | F | L | D | R | D | L | M | T | L | P | I | Q | N | K | L | A | P | F | I | A | K | R | R | T | P | K | I | Q | E | Q | Y | R | R | I | G | G | G | S | P | I | K | I | W | T | S | K | Q | G | E | G | M | V | K | L | L | D | E | L | S | P | N | T | A | P | H | K | Y | Y | I | G | F | R | Y | V | H | - | - | - | - | - | - | - | - | - | - | - | P | L | T | E | E | A | I | E | E | M | E | R | D | G | L | E | R | A | I | A | F | T | Q | Y | P | Q | Y | S | C | S | T | T | G | S | S | L | N | A | I | Y | R | Y | Y | N | Q | V | G | R | K | P | T | M | K | W | S | T | I | D | R | W | P | T | H | H | L | L | I | Q | C | F | A | D | H | I | L | K | E | L | - | D | H | F | P | L | E | K | R | S | E | V | V | I | L | F | S | A | H | S | L | P | M | S | V | V | N | R | G | D | P | Y | P | Q | E | V | S | A | T | V | Q | K | V | M | E | R | L | E | Y | C | N | P | Y | R | L | V | W | Q | S | K | - | V | G | P | M | P | W | L | G | P | Q | T | D | E | S | I | K | G | L | C | E | R | - | G | R | K | N | I | L | L | V | P | I | A | F | T | S | D | H | I | E | T | L | Y | E | L | D | I | - | - | - | - | E | Y | S | Q | V | L | A | K | E | C | G | - | - | - | - | - | - | - | - | - | - | - | - | - | V | E | N | I | R | R | A | E | S | L | N | G | N | P | L | F | S | K | A | L | A | D | L | V | H | S | H | I | Q | S | N | E | L | C | S | K | Q | L | T | L | S | C | P | L | C | V | N | P | V | C | R | E | T | K | S | F | F | T | S | Q | Q | L |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1qgoa_ | A | Q05592 | GO:0006779 |
| d1qgoa_ | A | Q05592 | GO:0009236 |
| d1qgoa_ | A | Q05592 | GO:0016829 |
| d1qgoa_ | A | Q05592 | GO:0016852 |
| d1qgoa_ | A | Q05592 | GO:0019251 |
| d1qgoa_ | A | Q05592 | GO:0046872 |
| d1qgoa_ | A | Q05592 | GO:0046906 |
| d1qgoa_ | A | Q05592 | GO:0050897 |
