solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1b74a1 ( 1 ) mkiGIFdsg--vgGl--tvlkairnryr--------kvdIvylg---dt
d1b74a2 ( 106 ) nkkiGVIGt--patvksgayqrkleeg----------gadVfAka--CPl
d1jfla1 ( 1 ) mktiGILGGMgplaTa--elfrrivikt--pAkrdqehpkViifn---np
d1jfla2 ( 116 ) fkkAGLLAt--tgtivsgvyekefsk-y---------gveImtPtedeqk
d2zska1 ( 3 ) kiGiIGGttpestl--yyykkyieiSrekfe-y-fypeLiiys---I-
d2zska2 ( 115 ) vrkVLLLGt---ttMtadfyiktlee-k---------gleVvvPndeek-
d5hqta1 ( 1 ) mktiGLLGGMswesti--pyyrlinegikqrlggl-hsAqVllhs---v-
d5hqta2 ( 117 ) mtrVALLGt--rytMeqdfyrgrlteqf---------sinCliPeadera
bbbb aaaaa aaaaaaaa bb
60 70 80 90 100
d1b74a1 ( 35 ) a-rvpYgir----s-kdtIIrySleCAgfLkdk-gVdiIvVAc-ntASay
d1b74a2 ( 142 ) faplaeeglLeg----eiTrkvVehyLkeFk--gkIdtLilgcthyPLl-
d1jfla1 ( 44 ) q-Ipdrtayilgkg--edPrpqLiwTAkrLeec-gAdfIiMPc-ntAHa-
d1jfla2 ( 154 ) dVmrGIyeGvkagn-lklGreLLlkTAkiLeer-gAeCIIAGc-teVsv-
d2zska1 ( 44 ) n-f-eFfqnp--eg-weGrkkiLinAAkaLera-gAelIaFAa-ntPHl-
d2zska2 ( 153 ) eLnrIIfeeLafgn-lk-nkewIvrLIekyresegIeGViLGc-teLpl-
d5hqta1 ( 44 ) d-fheIeeCqrrge-wdkTGdiLAeAAlglqra-gAegIvLCt-ntMHk-
d5hqta2 ( 156 ) kInqIIfeeLclgqfteasrayYaqVIarLaeq-gAqGViFGc-teIgl-
aaaaaaaaaaaaaa bbbb
110 120 130 140 150
d1b74a1 ( 77 ) AlerLkkeI-nvpVfGviepgvkealkks--r
d1b74a2 ( 185 ) -kkeIkkfLgdaevvdssealslslhnfikddgssslelfftdlspnlqf
d1jfla1 ( 88 ) fvedIrkaI-kipIiSmieetakkvkelg
d1jfla2 ( 200 ) VLk--qddL-kvplidpmdviaevavkvalek
d2zska1 ( 87 ) vfddVqreV-nvpMvsiidavaeeilkrg
d2zska2 ( 199 ) AI---qgdV-sVeVfdsaeihmrklielase
d5hqta1 ( 89 ) vAdaIesrC-tlpFlhiadatgraitgag
d5hqta2 ( 203 ) LVp--eerS-vlpVfdtaaihaedavafml-s
aaaaaaaaaaa
160
d1b74a1
d1b74a2 ( 234 ) liklilgrdypVklaegvf
d1jfla1
d1jfla2
d2zska1
d2zska2
d5hqta1
d5hqta2
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Glutamate racemase | Aspartate/glutamate racemase | Aquifex pyrophilus [TaxId: 2714] | 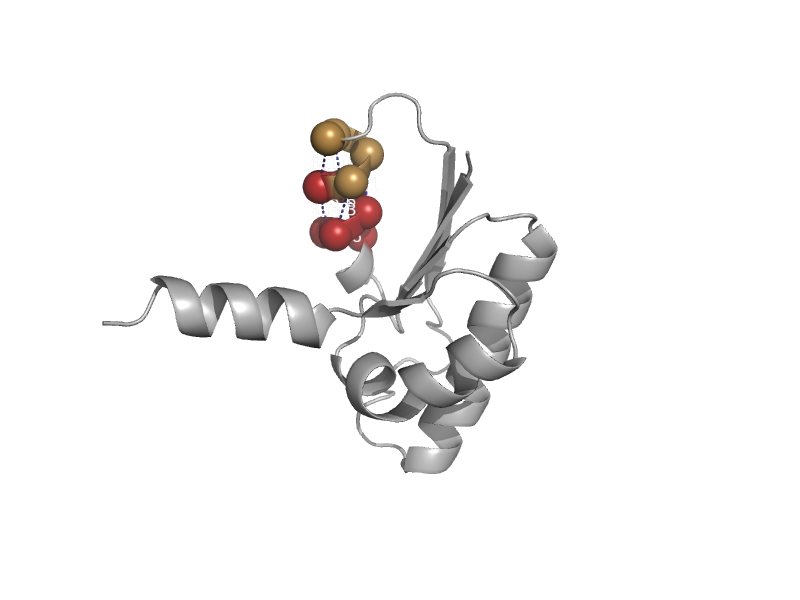 | view | |
| Glutamate racemase | Aspartate/glutamate racemase | Aquifex pyrophilus [TaxId: 2714] | 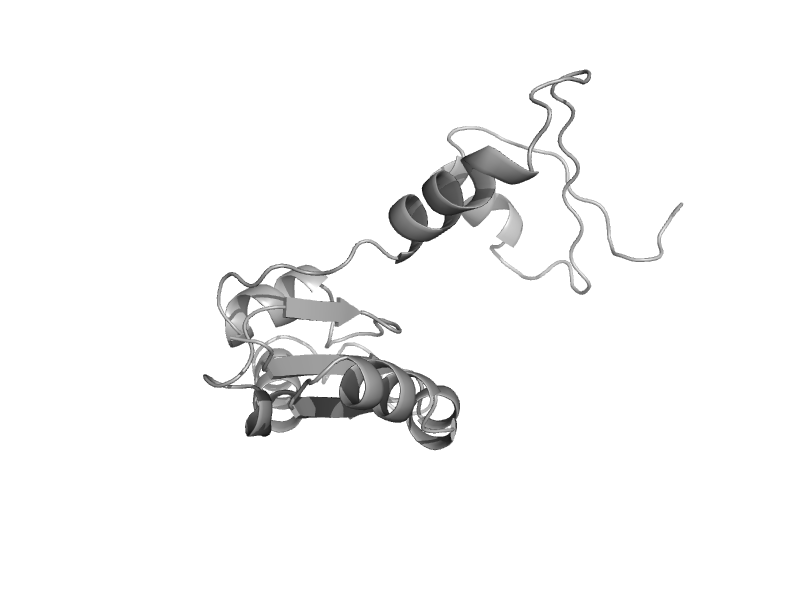 | view | |
| Aspartate racemase | Aspartate/glutamate racemase | Pyrococcus horikoshii OT3 [TaxId: 70601] | 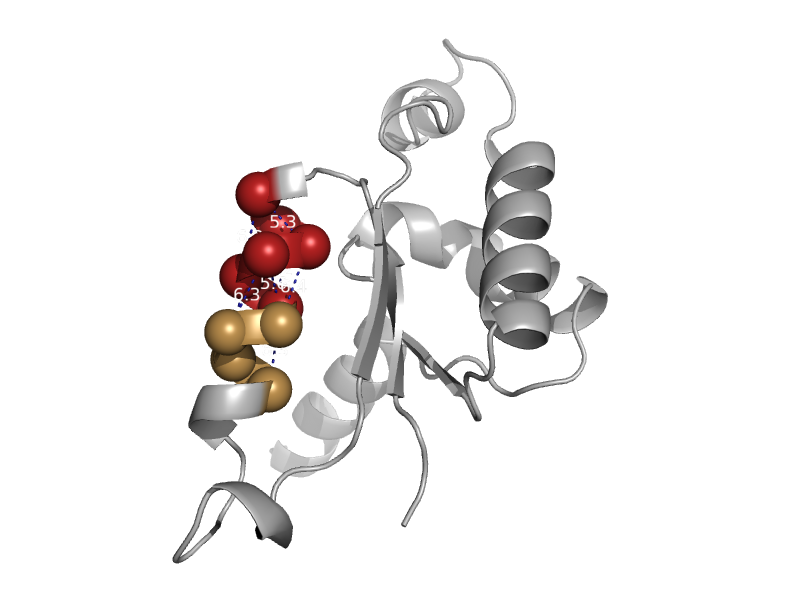 | view | |
| Aspartate racemase | Aspartate/glutamate racemase | Pyrococcus horikoshii OT3 [TaxId: 70601] | 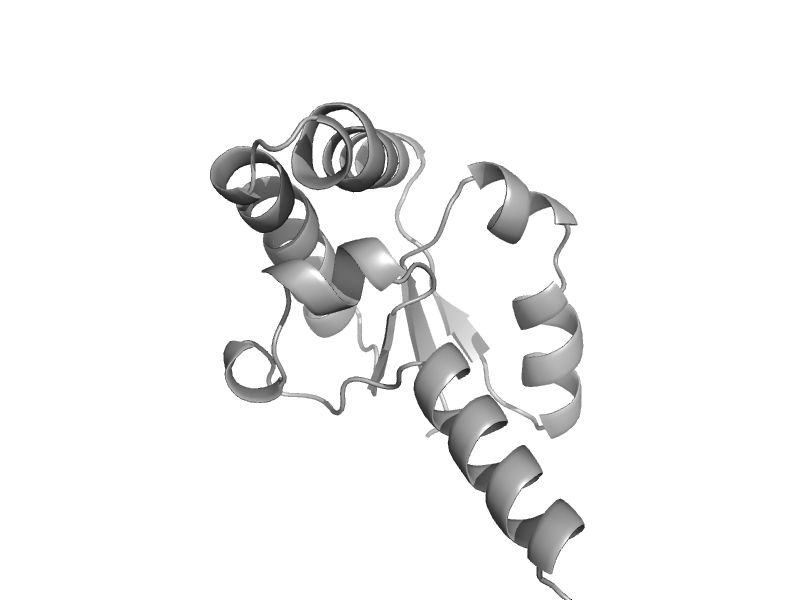 | view | |
| automated matches | automated matches | Pyrococcus horikoshii OT3 [TaxId: 70601] | 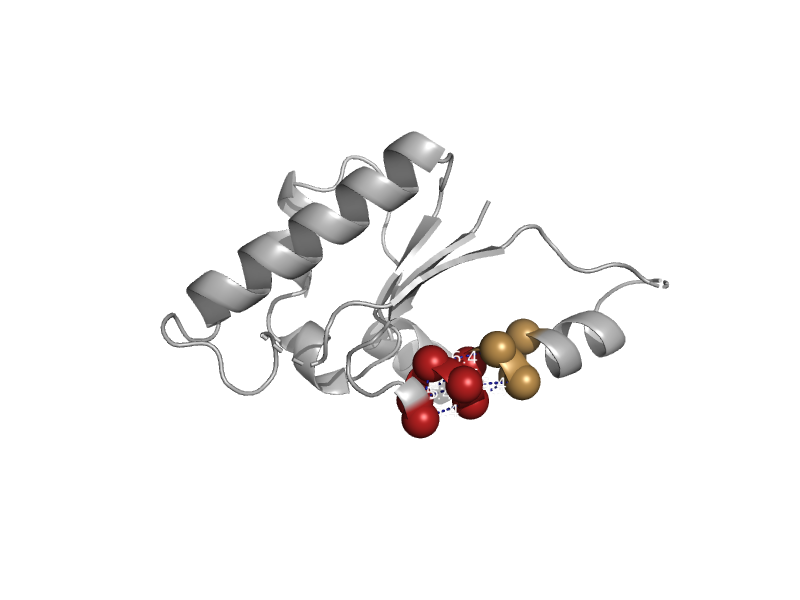 | view | |
| automated matches | automated matches | Pyrococcus horikoshii OT3 [TaxId: 70601] | 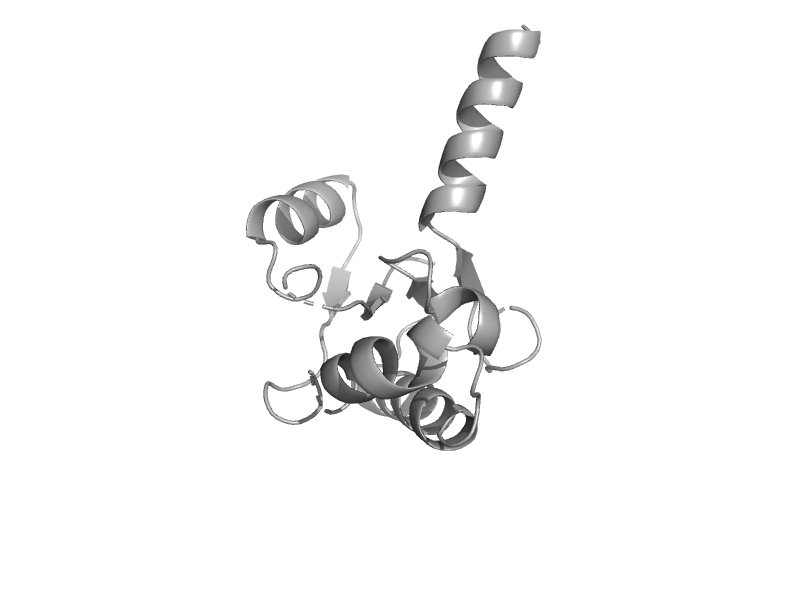 | view | |
| automated matches | automated matches | Escherichia coli [TaxId: 1330457] | 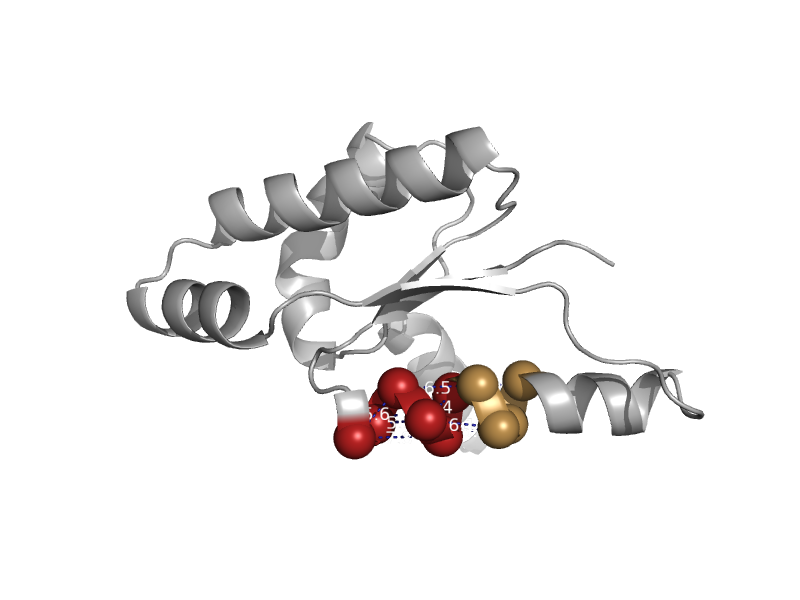 | view | |
| automated matches | automated matches | Escherichia coli [TaxId: 1330457] | 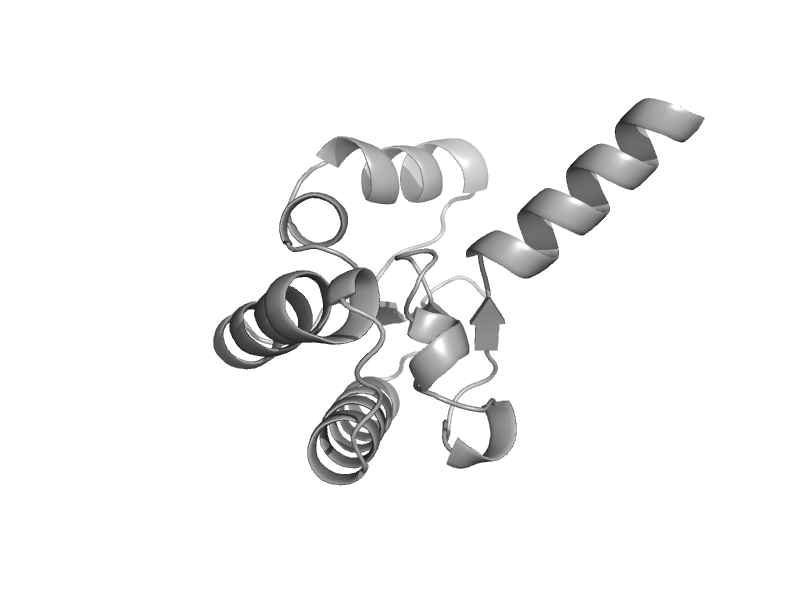 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | M | K | I | G | I | F | D | S | G | - | - | V | G | G | L | - | - | T | V | L | K | A | I | R | N | R | Y | R | - | - | - | - | - | - | - | - | K | V | D | I | V | Y | L | G | - | - | - | D | T | A | - | R | V | P | Y | G | I | R | - | - | - | - | S | - | K | D | T | I | I | R | Y | S | L | E | C | A | G | F | L | K | D | K | - | G | V | D | I | I | V | V | A | C | - | N | T | A | S | A | Y | A | L | E | R | L | K | K | E | I | - | N | V | P | V | F | G | V | I | E | P | G | V | K | E | A | L | K | K | S | - | - | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| N | K | K | I | G | V | I | G | T | - | - | P | A | T | V | K | S | G | A | Y | Q | R | K | L | E | E | G | - | - | - | - | - | - | - | - | - | - | G | A | D | V | F | A | K | A | - | - | C | P | L | F | A | P | L | A | E | E | G | L | L | E | G | - | - | - | - | E | I | T | R | K | V | V | E | H | Y | L | K | E | F | K | - | - | G | K | I | D | T | L | I | L | G | C | T | H | Y | P | L | L | - | - | K | K | E | I | K | K | F | L | G | D | A | E | V | V | D | S | S | E | A | L | S | L | S | L | H | N | F | I | K | D | D | G | S | S | S | L | E | L | F | F | T | D | L | S | P | N | L | Q | F | L | I | K | L | I | L | G | R | D | Y | P | V | K | L | A | E | G | V | F |
| M | K | T | I | G | I | L | G | G | M | G | P | L | A | T | A | - | - | E | L | F | R | R | I | V | I | K | T | - | - | P | A | K | R | D | Q | E | H | P | K | V | I | I | F | N | - | - | - | N | P | Q | - | I | P | D | R | T | A | Y | I | L | G | K | G | - | - | E | D | P | R | P | Q | L | I | W | T | A | K | R | L | E | E | C | - | G | A | D | F | I | I | M | P | C | - | N | T | A | H | A | - | F | V | E | D | I | R | K | A | I | - | K | I | P | I | I | S | M | I | E | E | T | A | K | K | V | K | E | L | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| F | K | K | A | G | L | L | A | T | - | - | T | G | T | I | V | S | G | V | Y | E | K | E | F | S | K | - | Y | - | - | - | - | - | - | - | - | - | G | V | E | I | M | T | P | T | E | D | E | Q | K | D | V | M | R | G | I | Y | E | G | V | K | A | G | N | - | L | K | L | G | R | E | L | L | L | K | T | A | K | I | L | E | E | R | - | G | A | E | C | I | I | A | G | C | - | T | E | V | S | V | - | V | L | K | - | - | Q | D | D | L | - | K | V | P | L | I | D | P | M | D | V | I | A | E | V | A | V | K | V | A | L | E | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | K | I | G | I | I | G | G | T | T | P | E | S | T | L | - | - | Y | Y | Y | K | K | Y | I | E | I | S | R | E | K | F | E | - | Y | - | F | Y | P | E | L | I | I | Y | S | - | - | - | I | - | N | - | F | - | E | F | F | Q | N | P | - | - | E | G | - | W | E | G | R | K | K | I | L | I | N | A | A | K | A | L | E | R | A | - | G | A | E | L | I | A | F | A | A | - | N | T | P | H | L | - | V | F | D | D | V | Q | R | E | V | - | N | V | P | M | V | S | I | I | D | A | V | A | E | E | I | L | K | R | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| V | R | K | V | L | L | L | G | T | - | - | - | T | T | M | T | A | D | F | Y | I | K | T | L | E | E | - | K | - | - | - | - | - | - | - | - | - | G | L | E | V | V | V | P | N | D | E | E | K | - | E | L | N | R | I | I | F | E | E | L | A | F | G | N | - | L | K | - | N | K | E | W | I | V | R | L | I | E | K | Y | R | E | S | E | G | I | E | G | V | I | L | G | C | - | T | E | L | P | L | - | A | I | - | - | - | Q | G | D | V | - | S | V | E | V | F | D | S | A | E | I | H | M | R | K | L | I | E | L | A | S | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M | K | T | I | G | L | L | G | G | M | S | W | E | S | T | I | - | - | P | Y | Y | R | L | I | N | E | G | I | K | Q | R | L | G | G | L | - | H | S | A | Q | V | L | L | H | S | - | - | - | V | - | D | - | F | H | E | I | E | E | C | Q | R | R | G | E | - | W | D | K | T | G | D | I | L | A | E | A | A | L | G | L | Q | R | A | - | G | A | E | G | I | V | L | C | T | - | N | T | M | H | K | - | V | A | D | A | I | E | S | R | C | - | T | L | P | F | L | H | I | A | D | A | T | G | R | A | I | T | G | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M | T | R | V | A | L | L | G | T | - | - | R | Y | T | M | E | Q | D | F | Y | R | G | R | L | T | E | Q | F | - | - | - | - | - | - | - | - | - | S | I | N | C | L | I | P | E | A | D | E | R | A | K | I | N | Q | I | I | F | E | E | L | C | L | G | Q | F | T | E | A | S | R | A | Y | Y | A | Q | V | I | A | R | L | A | E | Q | - | G | A | Q | G | V | I | F | G | C | - | T | E | I | G | L | - | L | V | P | - | - | E | E | R | S | - | V | L | P | V | F | D | T | A | A | I | H | A | E | D | A | V | A | F | M | L | - | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
