solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1bgva2 ( 1 ) skyVdrViaeVekkyadepeFvqTVeevLsslgpvVdahpeyee
d1c1da2 ( 1 )
d1gtma2 ( 3 ) adpyeiVikqleraaqy------m------------------ei
d1hwxa3 ( 1 ) adreddpnffkmVegffdrgasi------vedklvedlktrqtqeqkrnr
d3vpxa1 ( 1 )
60 70 80 90 100
d1bgva2 ( 45 ) -valLermV--iPerviefrVpWeddngkvhvntGyRVQFngaigpykgG
d1c1da2 ( 1 ) sidsal-nwdgemtvtrfda-------mtgAhFVIRLdstqlgpAAgG
d1gtma2 ( 23 ) seealeflk--rpqrivevtIpVemddgsvkvftGFRVQHnwargpTkgG
d1hwxa3 ( 45 ) Vrgilriik--pCnhvlslsFpirrddgswevIeGyRAQHshqrtpCkGG
d3vpxa1 ( 1 ) meifkymehqdYeqLvicqdk-------aSgLkAIIAIhdttlgpAlGG
aaaaa bbbbbbb bbbbbbbbb bb
110 120 130 140 150
d1bgva2 ( 92 ) LrFap----sVnlSiMKflGFeqafkDslttlpmgGAKGGSdF--dpngk
d1c1da2 ( 41 ) TrAaqysnladALtdAGklAgamtlkmavsnlpmGGGKSVIaLpaprhsI
d1gtma2 ( 71 ) IrWhp----eEtlstVkalAAwmtwkTavmdlpygGGkGGIiv--dpkkL
d1hwxa3 ( 93 ) IrYst----dVsvdeVkAlASlmTykcavvdvpfGGAkAGVkI--npkny
d3vpxa1 ( 43 ) TrMwtYaseeeAIedALrlArgmtykNaaaglnlgGGKTVIig--npktd
bbb aaaaaaaaaaaaaaaaa bbbbbbb
160 170 180 190 200
d1bgva2 ( 136 ) sdreVmrFCqaFMteLyr--hIgpdiDvPagdlgVgarEigyMygqYrki
d1c1da2 ( 91 ) dpstwarILrIHAenIdk----lsgnYwTgpdvnTnsadMdtLndtTe--
d1gtma2 ( 115 ) sdrekerLArgYIraIyd--vIsPyeDiPapdvyTnpqIMawMMdeYe-t
d1hwxa3 ( 137 ) tdeDLekITrrFTmeLAkkgfIgpgvDVPapnmsTgerEMswIAdtyast
d3vpxa1 ( 91 ) kn---deMFraFGryIeg----lngrYiTaedvgTteadmdlInleTd--
aaaaaaaaaaaaaaa bb aaaaaaaaaa
210 220
d1bgva2 ( 184 ) vg---gfyngVLtg
d1c1da2 ( 135 ) ----------fVfgrslerggags
d1gtma2 ( 162 ) isrrktpAfgIItgkplsi
d1hwxa3 ( 187 ) iGhydinAhaCVtgKpisqggi
d3vpxa1 ( 132 ) ----------yV
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Glutamate dehydrogenase | Aminoacid dehydrogenases | Clostridium symbiosum [TaxId: 1512] | 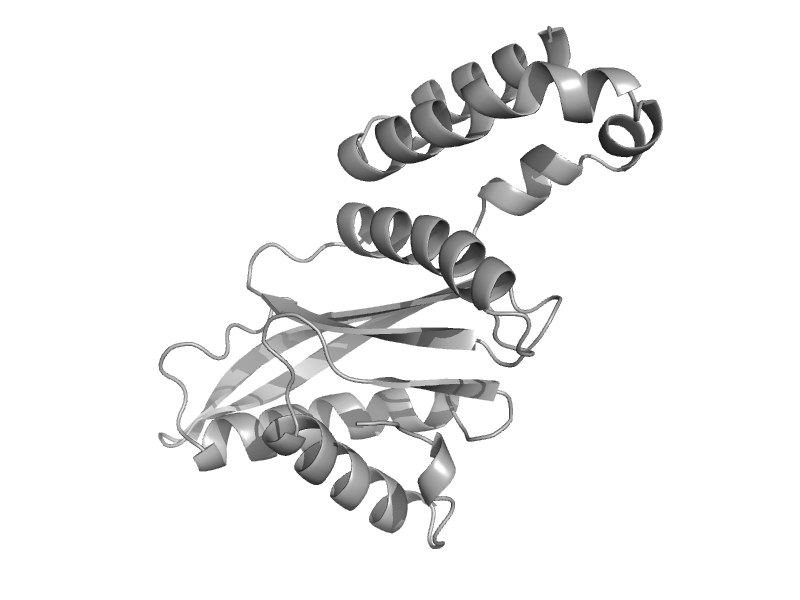 | view | |
| Phenylalanine dehydrogenase | Aminoacid dehydrogenases | Rhodococcus sp., M4 [TaxId: 1831] | 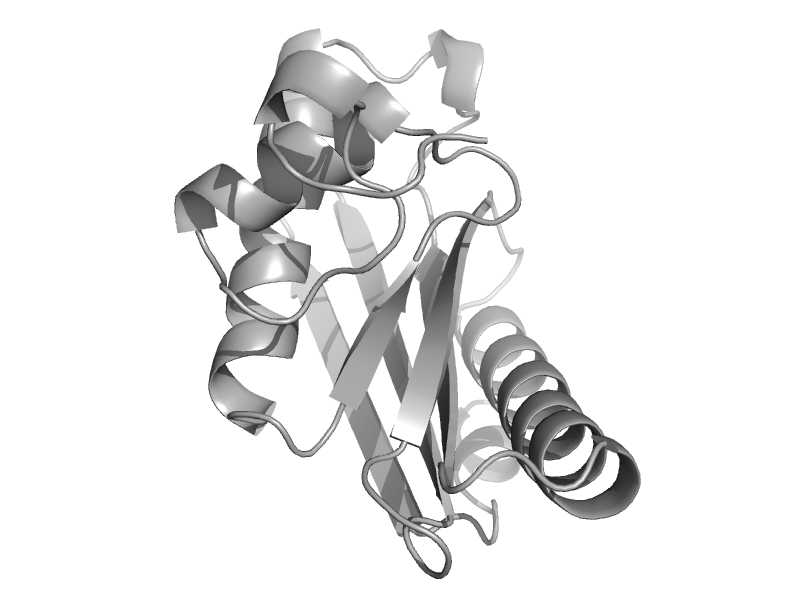 | view | |
| Glutamate dehydrogenase | Aminoacid dehydrogenases | Pyrococcus furiosus [TaxId: 2261] | 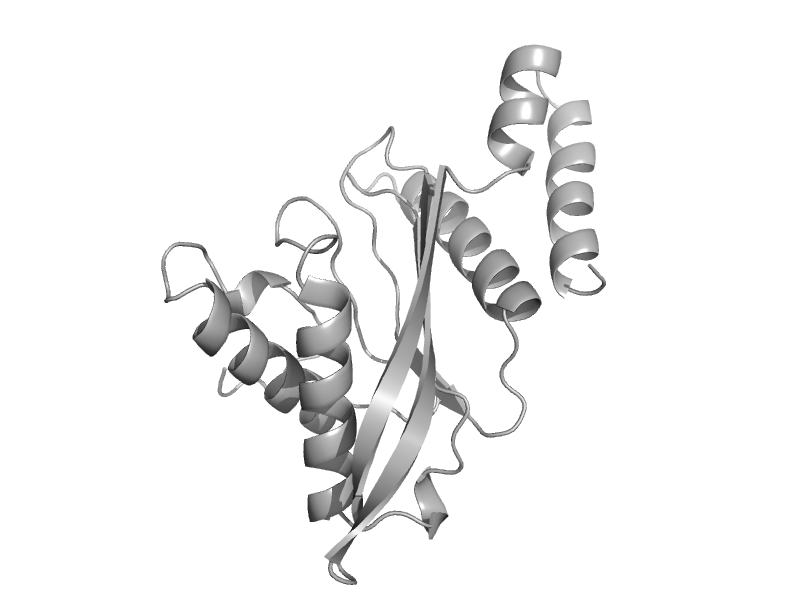 | view | |
| Glutamate dehydrogenase | Aminoacid dehydrogenases | Cow (Bos taurus) [TaxId: 9913] | 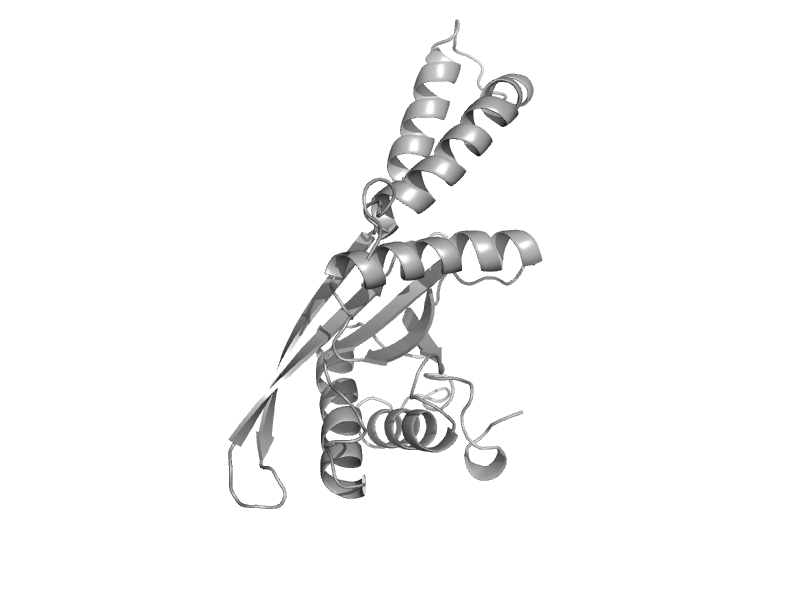 | view | |
| automated matches | automated matches | Sporosarcina psychrophila [TaxId: 1476] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | S | K | Y | V | D | R | V | I | A | E | V | E | K | K | Y | A | D | E | P | E | F | V | Q | T | V | E | E | V | L | S | S | L | G | P | V | V | D | A | H | P | E | Y | E | E | - | V | A | L | L | E | R | M | V | - | - | I | P | E | R | V | I | E | F | R | V | P | W | E | D | D | N | G | K | V | H | V | N | T | G | Y | R | V | Q | F | N | G | A | I | G | P | Y | K | G | G | L | R | F | A | P | - | - | - | - | S | V | N | L | S | I | M | K | F | L | G | F | E | Q | A | F | K | D | S | L | T | T | L | P | M | G | G | A | K | G | G | S | D | F | - | - | D | P | N | G | K | S | D | R | E | V | M | R | F | C | Q | A | F | M | T | E | L | Y | R | - | - | H | I | G | P | D | I | D | V | P | A | G | D | L | G | V | G | A | R | E | I | G | Y | M | Y | G | Q | Y | R | K | I | V | G | - | - | - | G | F | Y | N | G | V | L | T | G | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | I | D | S | A | L | - | N | W | D | G | E | M | T | V | T | R | F | D | A | - | - | - | - | - | - | - | M | T | G | A | H | F | V | I | R | L | D | S | T | Q | L | G | P | A | A | G | G | T | R | A | A | Q | Y | S | N | L | A | D | A | L | T | D | A | G | K | L | A | G | A | M | T | L | K | M | A | V | S | N | L | P | M | G | G | G | K | S | V | I | A | L | P | A | P | R | H | S | I | D | P | S | T | W | A | R | I | L | R | I | H | A | E | N | I | D | K | - | - | - | - | L | S | G | N | Y | W | T | G | P | D | V | N | T | N | S | A | D | M | D | T | L | N | D | T | T | E | - | - | - | - | - | - | - | - | - | - | - | - | F | V | F | G | R | S | L | E | R | G | G | A | G | S |
| - | - | - | - | - | - | A | D | P | Y | E | I | V | I | K | Q | L | E | R | A | A | Q | Y | - | - | - | - | - | - | M | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | I | S | E | E | A | L | E | F | L | K | - | - | R | P | Q | R | I | V | E | V | T | I | P | V | E | M | D | D | G | S | V | K | V | F | T | G | F | R | V | Q | H | N | W | A | R | G | P | T | K | G | G | I | R | W | H | P | - | - | - | - | E | E | T | L | S | T | V | K | A | L | A | A | W | M | T | W | K | T | A | V | M | D | L | P | Y | G | G | G | K | G | G | I | I | V | - | - | D | P | K | K | L | S | D | R | E | K | E | R | L | A | R | G | Y | I | R | A | I | Y | D | - | - | V | I | S | P | Y | E | D | I | P | A | P | D | V | Y | T | N | P | Q | I | M | A | W | M | M | D | E | Y | E | - | T | I | S | R | R | K | T | P | A | F | G | I | I | T | G | K | P | L | S | I | - | - | - | - | - |
| A | D | R | E | D | D | P | N | F | F | K | M | V | E | G | F | F | D | R | G | A | S | I | - | - | - | - | - | - | V | E | D | K | L | V | E | D | L | K | T | R | Q | T | Q | E | Q | K | R | N | R | V | R | G | I | L | R | I | I | K | - | - | P | C | N | H | V | L | S | L | S | F | P | I | R | R | D | D | G | S | W | E | V | I | E | G | Y | R | A | Q | H | S | H | Q | R | T | P | C | K | G | G | I | R | Y | S | T | - | - | - | - | D | V | S | V | D | E | V | K | A | L | A | S | L | M | T | Y | K | C | A | V | V | D | V | P | F | G | G | A | K | A | G | V | K | I | - | - | N | P | K | N | Y | T | D | E | D | L | E | K | I | T | R | R | F | T | M | E | L | A | K | K | G | F | I | G | P | G | V | D | V | P | A | P | N | M | S | T | G | E | R | E | M | S | W | I | A | D | T | Y | A | S | T | I | G | H | Y | D | I | N | A | H | A | C | V | T | G | K | P | I | S | Q | G | G | I | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | E | I | F | K | Y | M | E | H | Q | D | Y | E | Q | L | V | I | C | Q | D | K | - | - | - | - | - | - | - | A | S | G | L | K | A | I | I | A | I | H | D | T | T | L | G | P | A | L | G | G | T | R | M | W | T | Y | A | S | E | E | E | A | I | E | D | A | L | R | L | A | R | G | M | T | Y | K | N | A | A | A | G | L | N | L | G | G | G | K | T | V | I | I | G | - | - | N | P | K | T | D | K | N | - | - | - | D | E | M | F | R | A | F | G | R | Y | I | E | G | - | - | - | - | L | N | G | R | Y | I | T | A | E | D | V | G | T | T | E | A | D | M | D | L | I | N | L | E | T | D | - | - | - | - | - | - | - | - | - | - | - | - | Y | V | - | - | - | - | - | - | - | - | - | - | - | - |
