solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1a4ia2 ( 2 ) apaeilngkeisaqirarlknqvtqlkeqvpgftprlAILQVGnrddSn
d1edza2 ( 4 ) pgrtilaskvaetfnteiinnVeeykkthngqGpllVGFlANndpaak
d1luaa2 ( 2 ) skkllfQFDtd-----a
d1npya2 ( 1 ) minkdTqlcMsLsgrps--n-
d1nyta2 ( 1 ) metyAVFGnpia--hs
d3ngxa1 ( 2 ) kilrgeeiaekkaenlhgiiers-----glepslkLIQIgdneaAs
d3tnla1 ( 4 ) kiteritghTeliGLIAtpir--hs
d4b4ua1 ( 1 ) m--alvldgralakqieenllvrvealkakt-grtpilATIlVGddgaSa
d4cjxa1 ( 1 ) mpeavvidgravakaiqkelteevallerrykgrrpGlSTIICgkrkdsq
d5dzsa1 ( 1 ) mnffGlVGekLs--hs
d5o2aa1 ( 2 ) a-akiidgktiaqqvrseva-qvqariaagl-rapGlAvvlvgsnpasq
bbbbbb
60 70 80 90 100
d1a4ia2 ( 51 ) lyInvklkaAeeigikAthikLprtTteseVmkyItslne-----dstVh
d1edza2 ( 52 ) myAtwtqktsesmgfrYdlrvie---dkdfLeeaIiqAng-----ddsVn
d1luaa2 ( 14 ) tpsvfdvvvgydggadhItgyG--nVtpdnVgayVdgTIytrggkekqsT
d1npya2 ( 19 ) fgttfhnylydklglnfiYkaft--tq--diehaIkgVral------gir
d1nyta2 ( 15 ) kspfihqqfaqqlniehpYgrvl--ApindFintLnafFsa------gGk
d3ngxa1 ( 43 ) iyArakirrgkkigiaVdleky-ddismkdLlkrIddLak-----dpqin
d3tnla1 ( 27 ) lsptmhneafaklgldyvYlAfe--VgdkeLkdvVqgFram------nlr
d4b4ua1 ( 48 ) tyvrmkgnaCrrVgmdslkielpqeTtteqLlaeIekLna-----npdVh
d4cjxa1 ( 51 ) tyVrlkrkaaaaCgfrnfsvelpanVtqeaLereVirLne-----eeaCh
d5dzsa1 ( 15 ) vspqihkrvfeilnieSaYknfe--iskedIskldgaIkll------giq
d5o2aa1 ( 49 ) iyvaskrkaCeevgfvsrsydlpettseaellelidtlna-----dntid
aaaaaaaaaaa bbbbbb aaaaaaaaaa
110 120 130 140 150
d1a4ia2 ( 96 ) gFLVqlpLdSensInteeVinaIapekdvdg
d1edza2 ( 94 ) gImVYfpVfgna--qDqyLqqvVckekDveGlnhvYyqnlyhnvryldke
d1luaa2 ( 62 ) aIfVGGgdmaaGerVfeaVkkrFfgpfrVscmldSn
d1npya2 ( 59 ) gCaVsm-------pfketCmpfLd------eihpsAqaiesVntIvNdn-
d1nyta2 ( 57 ) gAnVtv-------pFkeeAfarAd------elterAalagaVntLmrled
d3ngxa1 ( 87 ) GIMIEnpLpk--gfdyyeIvrnIpyykdvda
d3tnla1 ( 69 ) gWnVsm-------pNktnIhkyLd------klspaAelvgaVntVvNdd-
d4b4ua1 ( 93 ) GILLqhpVp-a-qIderaCfdaIslakdvdg
d4cjxa1 ( 96 ) sIvVqlpLp-p-hIdkvaAlskIkpekdadc
d5dzsa1 ( 57 ) gVnVtv-------pYkerImkyLd------fiSpeAkrigaVntIllre-
d5o2aa1 ( 94 ) Gilvhlplp-a-gidnvkvlerihpdkdvdg
bbbb aaaa
d1a4ia2
d1edza2 ( 142 ) nrlkSi-l
d1luaa2
d1npya2 ( 95 ) gfLrAynt
d1nyta2 ( 94 ) grLlGdnt
d3ngxa1
d3tnla1 ( 105 ) gvLtGhit
d4b4ua1
d4cjxa1
d5dzsa1 ( 93 ) nmLyGynt
d5o2aa1
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Tetrahydrofolate dehydrogenase/cyclohydrolase | Tetrahydrofolate dehydrogenase/cyclohydrolase | Human (Homo sapiens) [TaxId: 9606] | 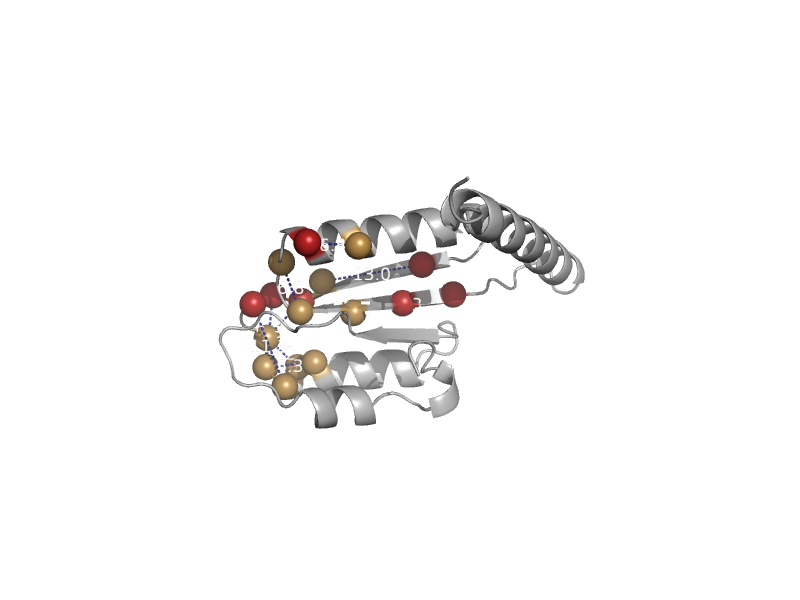 | view | |
| Tetrahydrofolate dehydrogenase/cyclohydrolase | Tetrahydrofolate dehydrogenase/cyclohydrolase | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 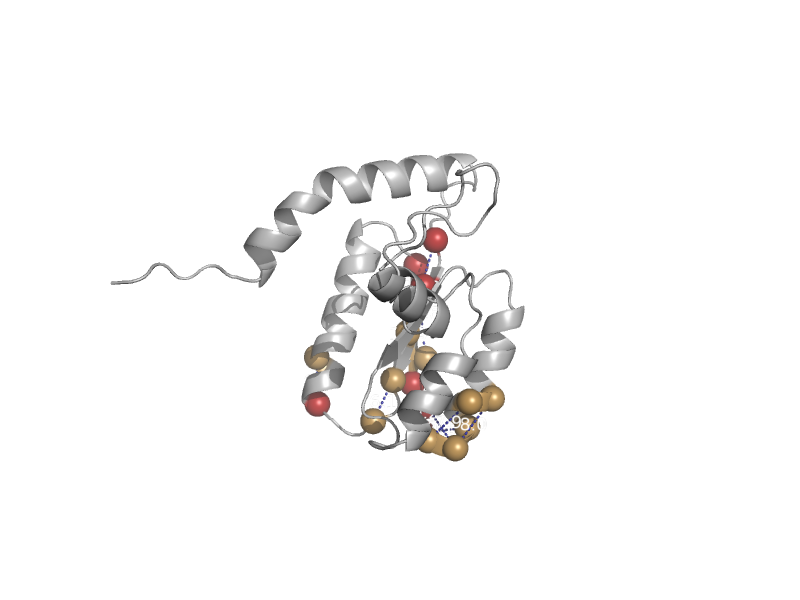 | view | |
| Methylene-tetrahydromethanopterin dehydrogenase | Methylene-tetrahydromethanopterin dehydrogenase | Methylobacterium extorquens [TaxId: 408] | 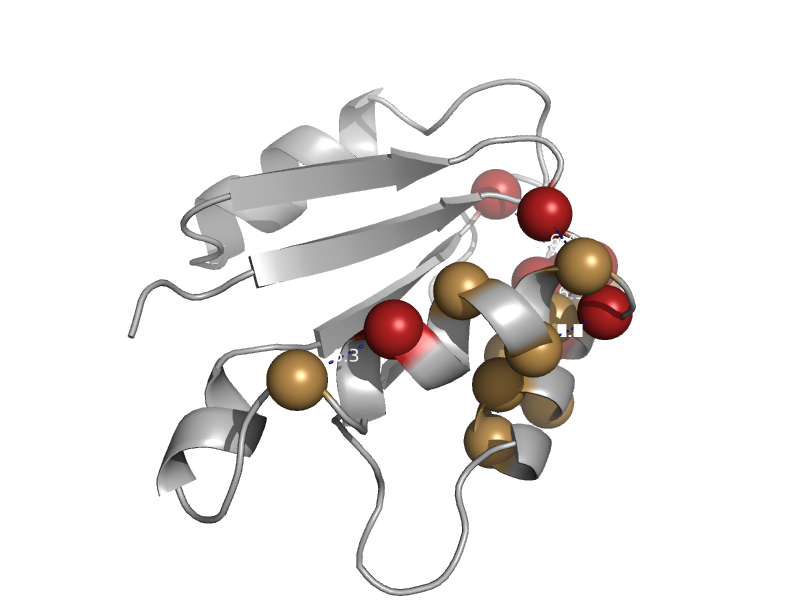 | view | |
| Shikimate 5-dehydrogenase-like protein HI0607 | Shikimate dehydrogenase-like | Haemophilus influenzae [TaxId: 727] | 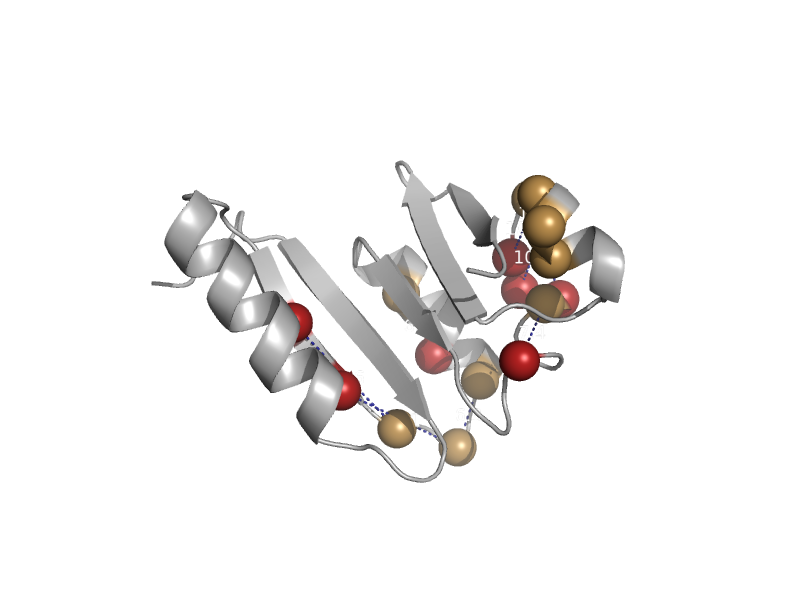 | view | |
| Shikimate 5-dehydrogenase AroE | Shikimate dehydrogenase-like | Escherichia coli [TaxId: 562] | 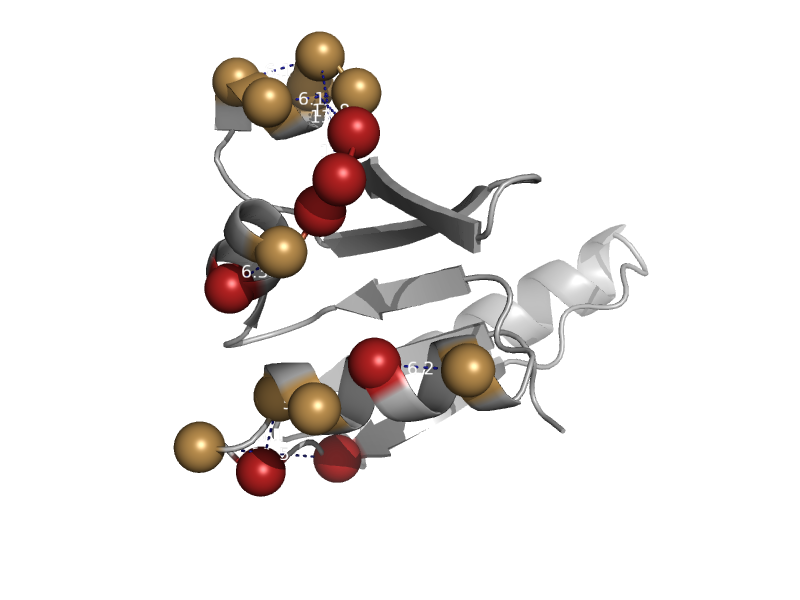 | view | |
| automated matches | automated matches | Thermoplasma acidophilum [TaxId: 2303] | 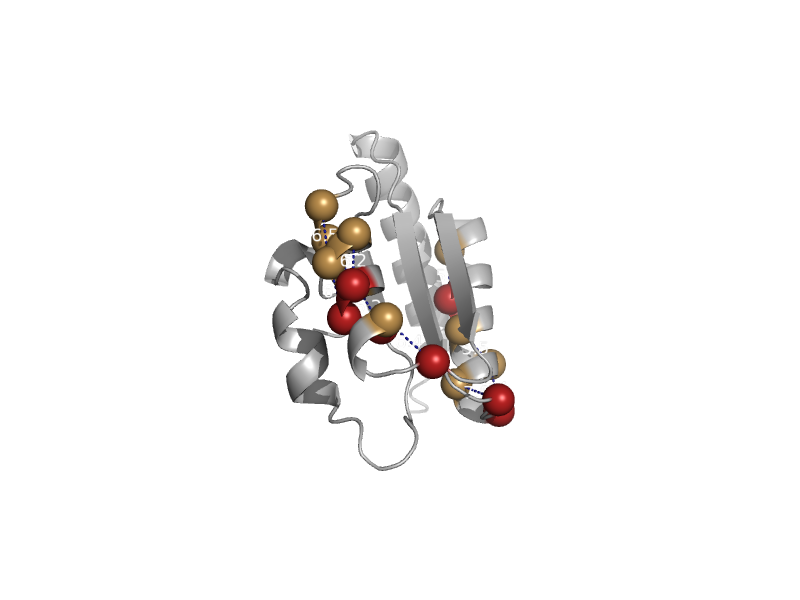 | view | |
| automated matches | automated matches | Listeria monocytogenes [TaxId: 169963] | 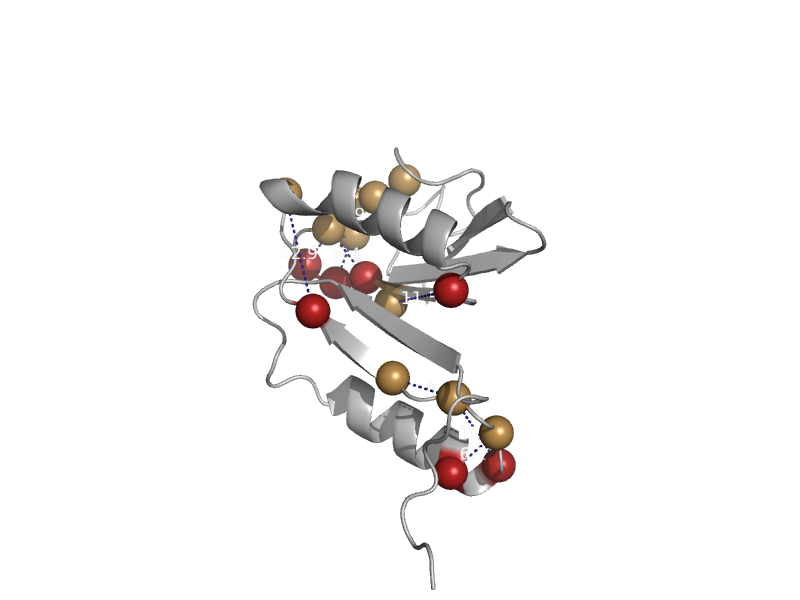 | view | |
| automated matches | automated matches | Acinetobacter baumannii [TaxId: 575584] | 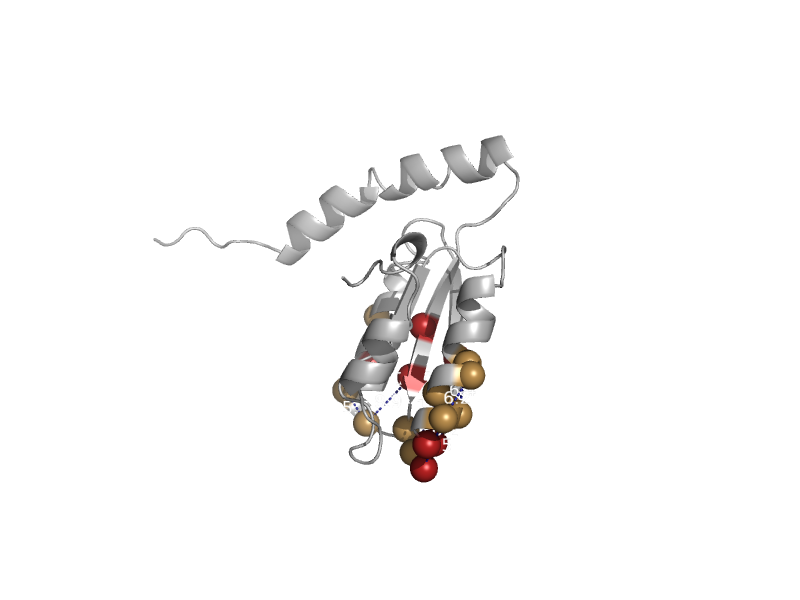 | view | |
| automated matches | automated matches | Trypanosoma brucei [TaxId: 999953] | 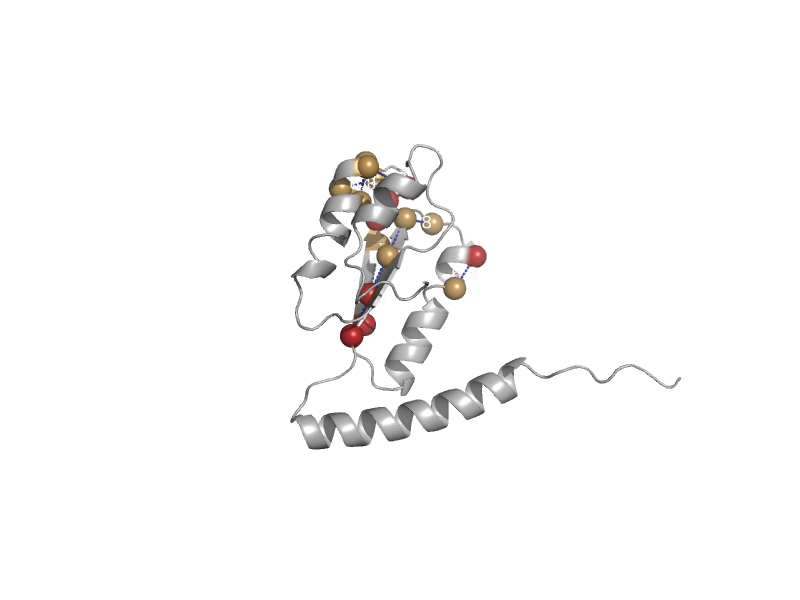 | view | |
| automated matches | automated matches | Peptoclostridium difficile [TaxId: 272563] | 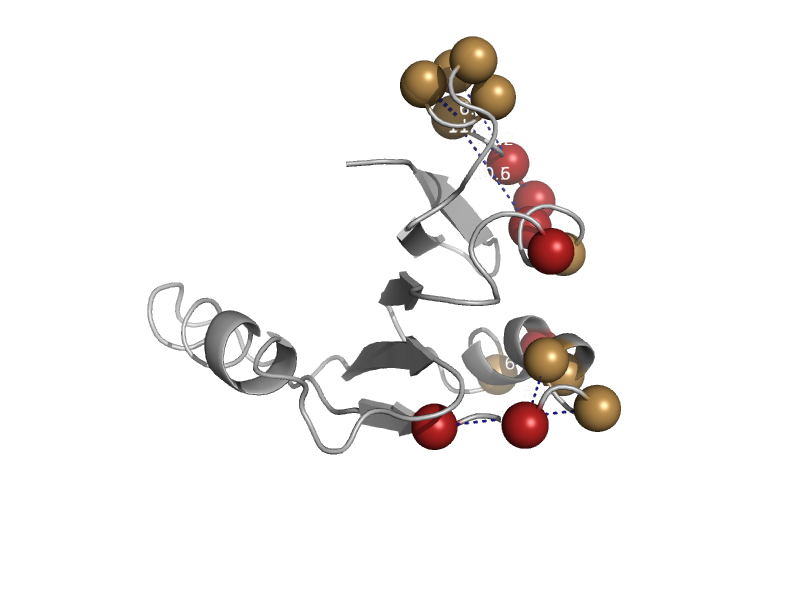 | view | |
| automated matches | Tetrahydrofolate dehydrogenase/cyclohydrolase | Escherichia coli [TaxId: 83333] | 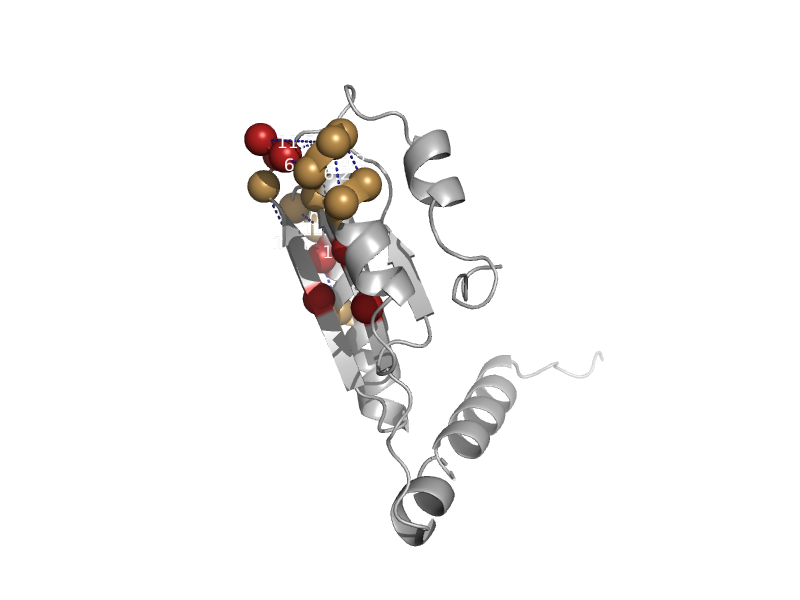 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | A | P | A | E | I | L | N | G | K | E | I | S | A | Q | I | R | A | R | L | K | N | Q | V | T | Q | L | K | E | Q | V | P | G | F | T | P | R | L | A | I | L | Q | V | G | N | R | D | D | S | N | L | Y | I | N | V | K | L | K | A | A | E | E | I | G | I | K | A | T | H | I | K | L | P | R | T | T | T | E | S | E | V | M | K | Y | I | T | S | L | N | E | - | - | - | - | - | D | S | T | V | H | G | F | L | V | Q | L | P | L | D | S | E | N | S | I | N | T | E | E | V | I | N | A | I | A | P | E | K | D | V | D | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | P | G | R | T | I | L | A | S | K | V | A | E | T | F | N | T | E | I | I | N | N | V | E | E | Y | K | K | T | H | N | G | Q | G | P | L | L | V | G | F | L | A | N | N | D | P | A | A | K | M | Y | A | T | W | T | Q | K | T | S | E | S | M | G | F | R | Y | D | L | R | V | I | E | - | - | - | D | K | D | F | L | E | E | A | I | I | Q | A | N | G | - | - | - | - | - | D | D | S | V | N | G | I | M | V | Y | F | P | V | F | G | N | A | - | - | Q | D | Q | Y | L | Q | Q | V | V | C | K | E | K | D | V | E | G | L | N | H | V | Y | Y | Q | N | L | Y | H | N | V | R | Y | L | D | K | E | N | R | L | K | S | I | - | L |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | K | K | L | L | F | Q | F | D | T | D | - | - | - | - | - | A | T | P | S | V | F | D | V | V | V | G | Y | D | G | G | A | D | H | I | T | G | Y | G | - | - | N | V | T | P | D | N | V | G | A | Y | V | D | G | T | I | Y | T | R | G | G | K | E | K | Q | S | T | A | I | F | V | G | G | G | D | M | A | A | G | E | R | V | F | E | A | V | K | K | R | F | F | G | P | F | R | V | S | C | M | L | D | S | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | I | N | K | D | T | Q | L | C | M | S | L | S | G | R | P | S | - | - | N | - | F | G | T | T | F | H | N | Y | L | Y | D | K | L | G | L | N | F | I | Y | K | A | F | T | - | - | T | Q | - | - | D | I | E | H | A | I | K | G | V | R | A | L | - | - | - | - | - | - | G | I | R | G | C | A | V | S | M | - | - | - | - | - | - | - | P | F | K | E | T | C | M | P | F | L | D | - | - | - | - | - | - | E | I | H | P | S | A | Q | A | I | E | S | V | N | T | I | V | N | D | N | - | G | F | L | R | A | Y | N | T |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | E | T | Y | A | V | F | G | N | P | I | A | - | - | H | S | K | S | P | F | I | H | Q | Q | F | A | Q | Q | L | N | I | E | H | P | Y | G | R | V | L | - | - | A | P | I | N | D | F | I | N | T | L | N | A | F | F | S | A | - | - | - | - | - | - | G | G | K | G | A | N | V | T | V | - | - | - | - | - | - | - | P | F | K | E | E | A | F | A | R | A | D | - | - | - | - | - | - | E | L | T | E | R | A | A | L | A | G | A | V | N | T | L | M | R | L | E | D | G | R | L | L | G | D | N | T |
| - | - | - | - | K | I | L | R | G | E | E | I | A | E | K | K | A | E | N | L | H | G | I | I | E | R | S | - | - | - | - | - | G | L | E | P | S | L | K | L | I | Q | I | G | D | N | E | A | A | S | I | Y | A | R | A | K | I | R | R | G | K | K | I | G | I | A | V | D | L | E | K | Y | - | D | D | I | S | M | K | D | L | L | K | R | I | D | D | L | A | K | - | - | - | - | - | D | P | Q | I | N | G | I | M | I | E | N | P | L | P | K | - | - | G | F | D | Y | Y | E | I | V | R | N | I | P | Y | Y | K | D | V | D | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | I | T | E | R | I | T | G | H | T | E | L | I | G | L | I | A | T | P | I | R | - | - | H | S | L | S | P | T | M | H | N | E | A | F | A | K | L | G | L | D | Y | V | Y | L | A | F | E | - | - | V | G | D | K | E | L | K | D | V | V | Q | G | F | R | A | M | - | - | - | - | - | - | N | L | R | G | W | N | V | S | M | - | - | - | - | - | - | - | P | N | K | T | N | I | H | K | Y | L | D | - | - | - | - | - | - | K | L | S | P | A | A | E | L | V | G | A | V | N | T | V | V | N | D | D | - | G | V | L | T | G | H | I | T |
| M | - | - | A | L | V | L | D | G | R | A | L | A | K | Q | I | E | E | N | L | L | V | R | V | E | A | L | K | A | K | T | - | G | R | T | P | I | L | A | T | I | L | V | G | D | D | G | A | S | A | T | Y | V | R | M | K | G | N | A | C | R | R | V | G | M | D | S | L | K | I | E | L | P | Q | E | T | T | T | E | Q | L | L | A | E | I | E | K | L | N | A | - | - | - | - | - | N | P | D | V | H | G | I | L | L | Q | H | P | V | P | - | A | - | Q | I | D | E | R | A | C | F | D | A | I | S | L | A | K | D | V | D | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| M | P | E | A | V | V | I | D | G | R | A | V | A | K | A | I | Q | K | E | L | T | E | E | V | A | L | L | E | R | R | Y | K | G | R | R | P | G | L | S | T | I | I | C | G | K | R | K | D | S | Q | T | Y | V | R | L | K | R | K | A | A | A | A | C | G | F | R | N | F | S | V | E | L | P | A | N | V | T | Q | E | A | L | E | R | E | V | I | R | L | N | E | - | - | - | - | - | E | E | A | C | H | S | I | V | V | Q | L | P | L | P | - | P | - | H | I | D | K | V | A | A | L | S | K | I | K | P | E | K | D | A | D | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | N | F | F | G | L | V | G | E | K | L | S | - | - | H | S | V | S | P | Q | I | H | K | R | V | F | E | I | L | N | I | E | S | A | Y | K | N | F | E | - | - | I | S | K | E | D | I | S | K | L | D | G | A | I | K | L | L | - | - | - | - | - | - | G | I | Q | G | V | N | V | T | V | - | - | - | - | - | - | - | P | Y | K | E | R | I | M | K | Y | L | D | - | - | - | - | - | - | F | I | S | P | E | A | K | R | I | G | A | V | N | T | I | L | L | R | E | - | N | M | L | Y | G | Y | N | T |
| - | A | - | A | K | I | I | D | G | K | T | I | A | Q | Q | V | R | S | E | V | A | - | Q | V | Q | A | R | I | A | A | G | L | - | R | A | P | G | L | A | V | V | L | V | G | S | N | P | A | S | Q | I | Y | V | A | S | K | R | K | A | C | E | E | V | G | F | V | S | R | S | Y | D | L | P | E | T | T | S | E | A | E | L | L | E | L | I | D | T | L | N | A | - | - | - | - | - | D | N | T | I | D | G | I | L | V | H | L | P | L | P | - | A | - | G | I | D | N | V | K | V | L | E | R | I | H | P | D | K | D | V | D | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
