solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1chma1 ( 2 ) qmpktlrirngdkvrstfsaqeYanrqarLrahLaaenIdAAIFtS----
d1pv9a1 ( 8 ) lvkfMdensIdrVFIak----
d1wl6a1 ( 1 ) seisrqefqrrRqaLVeqMqp-gSAALIFAapevt
aaa bbbb
60 70 80 90 100
d1chma1 ( 48 ) -----------yhNInyYSDFlYcsfgrpYALVVte-----ddVISISAn
d1pv9a1 ( 25 ) -----------pvNVyyfSgTspl---ggGyIIVdg-----deatLyVpe
d1wl6a1 ( 35 ) rsadseypyrQnsDFwyFTGfn----epeAVLVLIksddthnhsVLFNrv
aaaaaaa bbbb bbbbb
110 120 130 140 150
d1chma1 ( 82 ) id------------gGqPwrrTvgtdNIVYTdwqrdNYFaAIqqAlpkAr
d1pv9a1 ( 56 ) le------------yemAkeeSkl-pVvkfk------kfdeIYeiLknTe
d1wl6a1 ( 81 ) rdltaeiwfgrrlGqdaApeklgVdrAla-----fseInqqLyqlLngld
aaaaa bbb aaaaa
160 170 180 190 200
d1chma1 ( 120 ) rIGIEhdhLnlqnrdkLaarYpd------------AelvdVaaaCmrmr
d1pv9a1 ( 87 ) tLGiEgt-lsysmvenFkekSVk-------------efkkIddvikdlri
d1wl6a1 ( 126 ) VVyHaqgey-ayAdviVnsALekLrkgsrqnltaPatmidWrpvVhemrl
bbb aaaaaaaaa bbb aaaaaaa
d1chma1
d1pv9a1 ( 124 ) i
d1wl6a1 ( 175 ) fk
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Creatinase | Creatinase/prolidase N-terminal domain | Pseudomonas putida [TaxId: 303] | 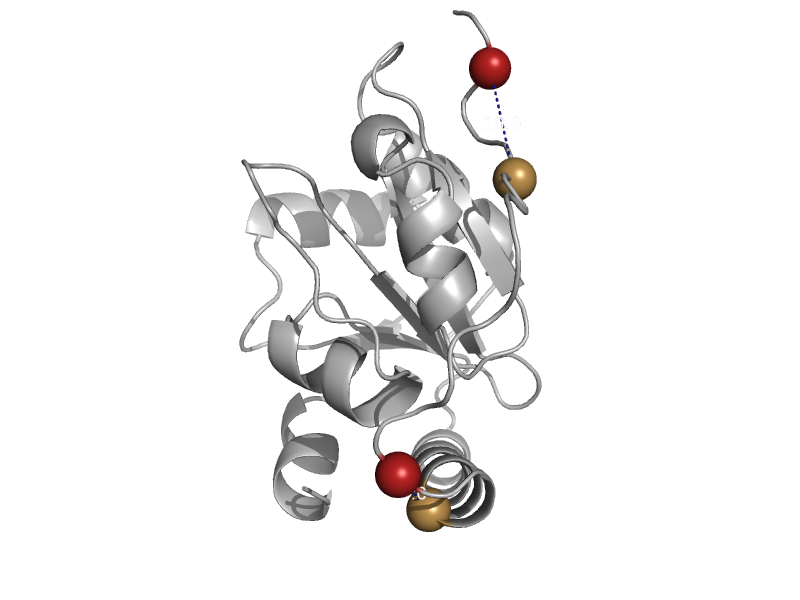 | view | |
| Aminopeptidase P | Creatinase/prolidase N-terminal domain | Pyrococcus furiosus [TaxId: 2261] | 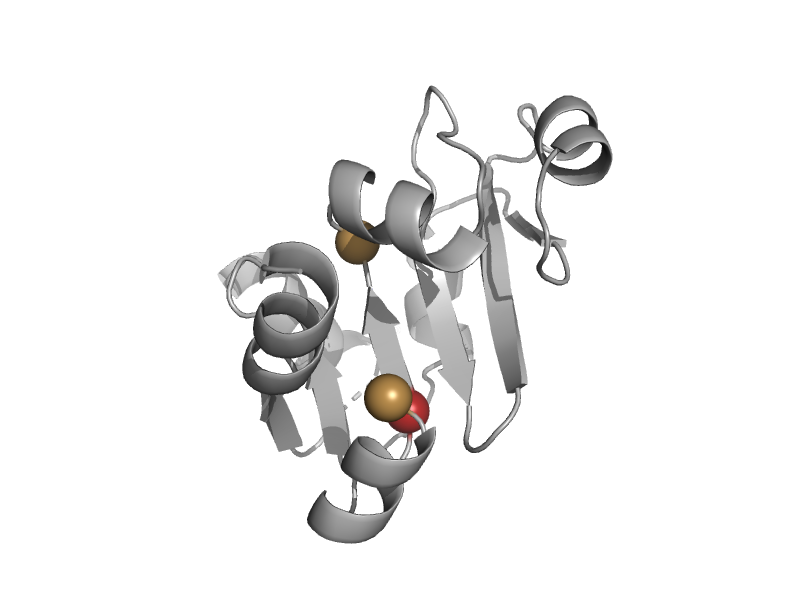 | view | |
| Aminopeptidase P | Creatinase/prolidase N-terminal domain | Escherichia coli [TaxId: 562] | 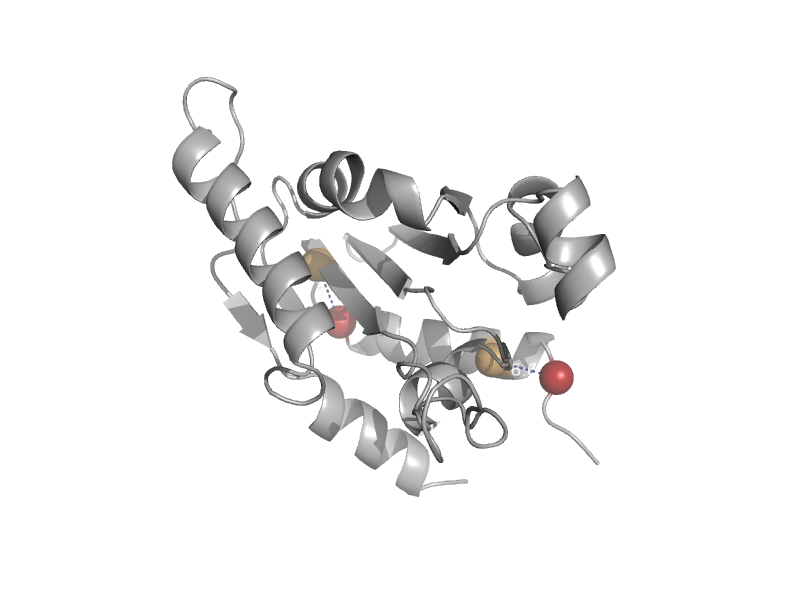 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q | M | P | K | T | L | R | I | R | N | G | D | K | V | R | S | T | F | S | A | Q | E | Y | A | N | R | Q | A | R | L | R | A | H | L | A | A | E | N | I | D | A | A | I | F | T | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | H | N | I | N | Y | Y | S | D | F | L | Y | C | S | F | G | R | P | Y | A | L | V | V | T | E | - | - | - | - | - | D | D | V | I | S | I | S | A | N | I | D | - | - | - | - | - | - | - | - | - | - | - | - | G | G | Q | P | W | R | R | T | V | G | T | D | N | I | V | Y | T | D | W | Q | R | D | N | Y | F | A | A | I | Q | Q | A | L | P | K | A | R | R | I | G | I | E | H | D | H | L | N | L | Q | N | R | D | K | L | A | A | R | Y | P | D | - | - | - | - | - | - | - | - | - | - | - | - | A | E | L | V | D | V | A | A | A | C | M | R | M | R | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | V | K | F | M | D | E | N | S | I | D | R | V | F | I | A | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | V | N | V | Y | Y | F | S | G | T | S | P | L | - | - | - | G | G | G | Y | I | I | V | D | G | - | - | - | - | - | D | E | A | T | L | Y | V | P | E | L | E | - | - | - | - | - | - | - | - | - | - | - | - | Y | E | M | A | K | E | E | S | K | L | - | P | V | V | K | F | K | - | - | - | - | - | - | K | F | D | E | I | Y | E | I | L | K | N | T | E | T | L | G | I | E | G | T | - | L | S | Y | S | M | V | E | N | F | K | E | K | S | V | K | - | - | - | - | - | - | - | - | - | - | - | - | - | E | F | K | K | I | D | D | V | I | K | D | L | R | I | I | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | E | I | S | R | Q | E | F | Q | R | R | R | Q | A | L | V | E | Q | M | Q | P | - | G | S | A | A | L | I | F | A | A | P | E | V | T | R | S | A | D | S | E | Y | P | Y | R | Q | N | S | D | F | W | Y | F | T | G | F | N | - | - | - | - | E | P | E | A | V | L | V | L | I | K | S | D | D | T | H | N | H | S | V | L | F | N | R | V | R | D | L | T | A | E | I | W | F | G | R | R | L | G | Q | D | A | A | P | E | K | L | G | V | D | R | A | L | A | - | - | - | - | - | F | S | E | I | N | Q | Q | L | Y | Q | L | L | N | G | L | D | V | V | Y | H | A | Q | G | E | Y | - | A | Y | A | D | V | I | V | N | S | A | L | E | K | L | R | K | G | S | R | Q | N | L | T | A | P | A | T | M | I | D | W | R | P | V | V | H | E | M | R | L | F | K |
