solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1ex2a- ( 1 ) mtkplILAsqsprrkeLLdllq-----l-pysiivs---eveekl
d1k7ka1 ( 2 ) qkVVlAtgnvgkvreLaslLd--fgl-dIvaqtdlgVdsaeet
d1u14a1 ( 2 ) hqVISAttnpakiqAIlqAFeeifgegsChItpv---aveSgv
d1v7ra- ( 1 ) mkIfFItsnpgkvrEVanfLgt-fgI-eIvqlk---heypeiq
d1vp2a- ( 3 ) kltVyLAttnphkv-eIkmIap--w-M-eIlpsp-----ivved
d2amha1 ( 7 ) eeirtMiIGTssafrAnVLrehFgdr-Fr-nfvllpp---dideka
d2cara1 ( 1 ) maasLvgkkIVFvtgnakkleeVvqiLgdkFPC-tLvaqk---idLpeyq
d4jhca- ( 2 ) pkLILAstspwrralLeklq-----i-sFecaap----vdetp
d4oo0a- ( 8 ) aptplfqtLYLAs-sprrqeLLqqig-----V--felllprpdedaala
bbb aaaaaaaa bb
60 70 80 90 100
d1ex2a- ( 37 ) nrnfspeenVqwlAkqkAkaVadlhp-----hAiVIGAdtmvÇLd---ge
d1k7ka1 ( 43 ) gl------tfieNAilkArhAAkvta------lpAIAddsgLaVdvlgga
d1u14a1 ( 42 ) peqpfgseeTraGArnrVdnArrlhp-----qAdfWVAIEaGIdd-----
d1v7ra- ( 39 ) ae------kLedVVdfGIswLkgkVp------epFMIEdsGLfIeslkgf
d1vp2a- ( 42 ) ge------tfleNSvkkAvvyGk-lk------hpVMADdSgLvIyslggf
d2amha1 ( 48 ) yraadpfelTesIArakMkaVlekarqh---paIALTFDqVVVkg---de
d2cara1 ( 47 ) g-------epdeISiqkCqeAvrqvq------gpVLVedtCLCFnalggl
d4jhca- ( 36 ) r-desprqLVlrLAqekAqsLasryp-----dhlIIGSDqvCvLd---ge
d4oo0a- ( 56 ) lpgeaadayVrrVTvakAeaArarLvasgkpaaPVLVAdtTVTId---ga
aaaaaaaaaaaaa bbbbbbbbbb
110 120 130 140 150
d1ex2a- ( 79 ) çLg-----kPq----dqeeAasMLrrLSg-----rsHsVITAVSIqA-en
d1k7ka1 ( 81 ) P-giysarysgedatdqkNLqkLletk-dvpddqrqArfhCVLVyLrhae
d1u14a1 ( 82 ) ------------------------------------datFSwVVIdngv-
d1v7ra- ( 77 ) P-gvyssyvyr-----tiglegIlklMega--edrrAyfkSVIGFyidg-
d1vp2a- ( 80 ) P-gvmsa-fm-eehsy-eKMrtIlkmLegkd---rrAAfVCSATFfdPve
d2amha1 ( 98 ) vre-----kPl----steqCrsfIasYsg-----ggVrTVATYALCvvgt
d2cara1 ( 84 ) PG-pyIkwfle-----klkpegLhqlLagf--edksAyALCTFALStGdp
d4jhca- ( 78 ) itg-----kPl----teenArlqLrkAsG-----niVtfyTGLALfnSan
d4oo0a- ( 103 ) iLg-----kPt----daddAlamLtrLag-----reHaVlTAVAVIdasg
a aaaaaaa bbbbbbbbbbb
160 170 180 190 200
d1ex2a- ( 114 ) --hsetfydkTeVaFwsLseeeIwtYIetkePmdkaGAYgIq----g-rG
d1k7ka1 ( 130 ) DptplvChGsWpGvItr----epagt----ggfgYDpIFfVp---segkT
d1u14a1 ( 95 ) ---qrGearSatlpLp---avIldrVrqgealgpVsqytgideigrkeGa
d1v7ra- ( 118 ) --kaykfsgvtwgrIsn----ekrgt----hgfgYDpIFIPe---gsekT
d1vp2a- ( 125 ) -ntlisvedrVeGrIAn----eirgt----ggfgYDpFFiPd---gydkT
d2amha1 ( 134 ) e-nvlvahneTeTfFskFgddiVerTlergaCmnsaGGLVVe----deDM
d2cara1 ( 126 ) sqpVrLFrgrTsGrIv-----aprgc----qdfgwdPCFqPd---gyeqT
d4jhca- ( 114 ) g-hlqtevEpFdVhFrhLseaeIdnYVrkehPlhcaGSFks-----g-fG
d4oo0a- ( 139 ) e-llppaLSrSsVrFaaasrdaYvrYvetgePfgkaGAYaIq----g-rA
bbbbbbbbbbb
210 220 230
d1ex2a- ( 157 ) alFvkkidg----dyySVMGLPiskTmralrhF
d1k7ka1 ( 169 ) AAeL----t-eeksaiShrgqAl-lLldalrn-g
d1u14a1 ( 140 ) iGvft----agkltrssvYyqAVilALspFhna
d1v7ra- ( 155 ) FAeM----tieeKNalSHRGkALkaFfewLkvnlky
d1vp2a- ( 163 ) FGei----p-hlKekiSHRSkAFrkLFsvLekil
d2amha1 ( 179 ) srhvvrivg----tsyGVrgMepavVekLLsql
d2cara1 ( 164 ) YAeM----pkaeknavShrfrALleLqeyFgs-laa
d4jhca- ( 158 ) itLFerlegr---DpnTLvgLPLiALcqMLrregnplm
d4oo0a- ( 183 ) aeFIeridg----shsGIMGLPlfeTaalLrtArVaf
aa aa aaaaaaaa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Maf protein | Maf-like | Bacillus subtilis [TaxId: 1423] | 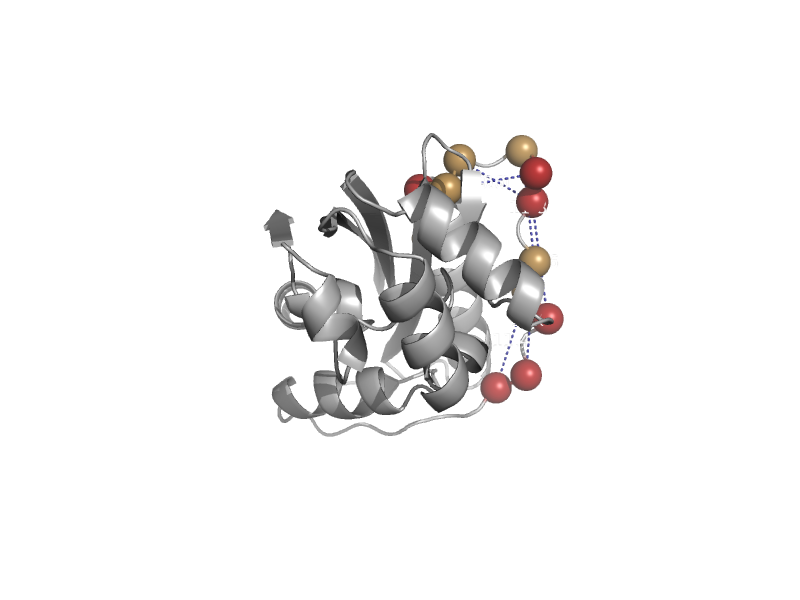 | view | |
| Hypothetical protein YggV | ITPase (Ham1) | Escherichia coli [TaxId: 562] | 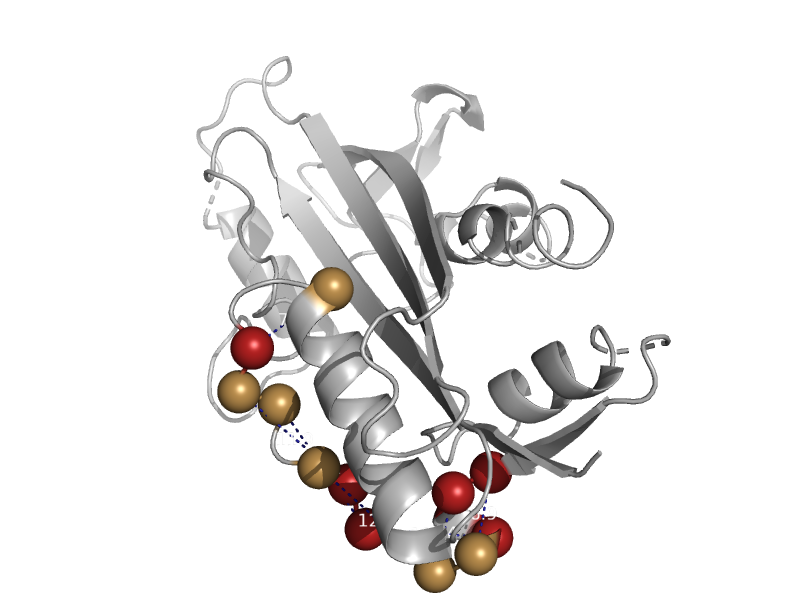 | view | |
| Hypothetical protein YjjX | YjjX-like | Salmonella typhimurium [TaxId: 90371] | 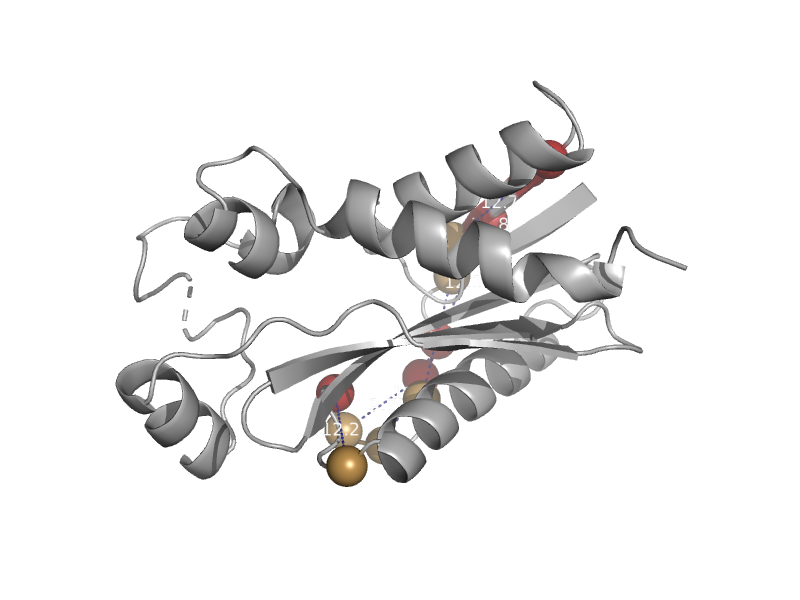 | view | |
| XTP pyrophosphatase | ITPase (Ham1) | Pyrococcus horikoshii [TaxId: 53953] | 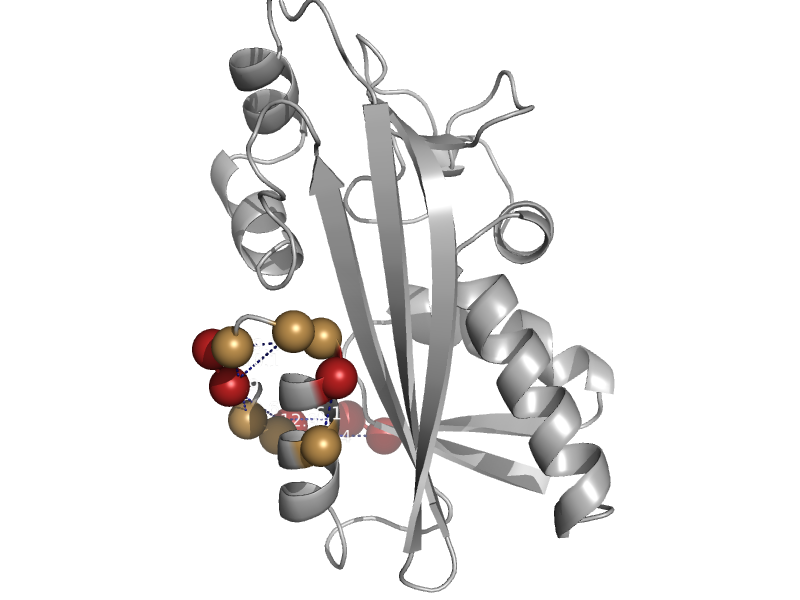 | view | |
| Putative inosine/xanthosine triphosphate pyrophosphatase TM0159 | ITPase (Ham1) | Thermotoga maritima [TaxId: 2336] | 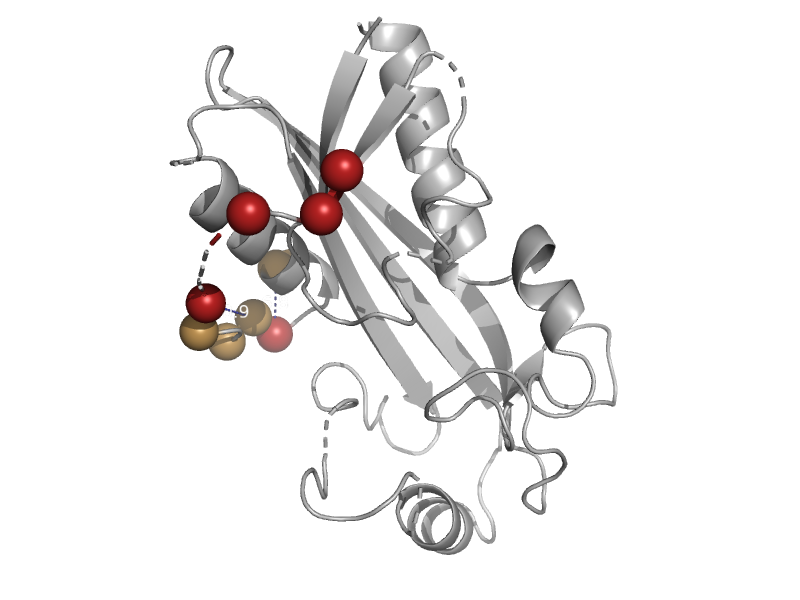 | view | |
| Maf homologue Tb11.01.5890 | Maf-like | Trypanosome (Trypanosoma brucei) [TaxId: 5691] | 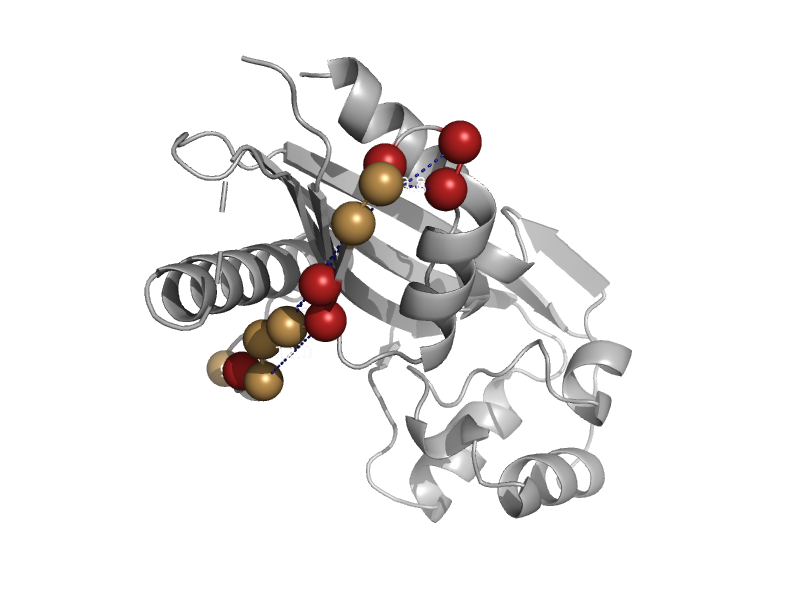 | view | |
| Inosine triphosphate pyrophosphatase, ITPase | ITPase (Ham1) | Human (Homo sapiens) [TaxId: 9606] | 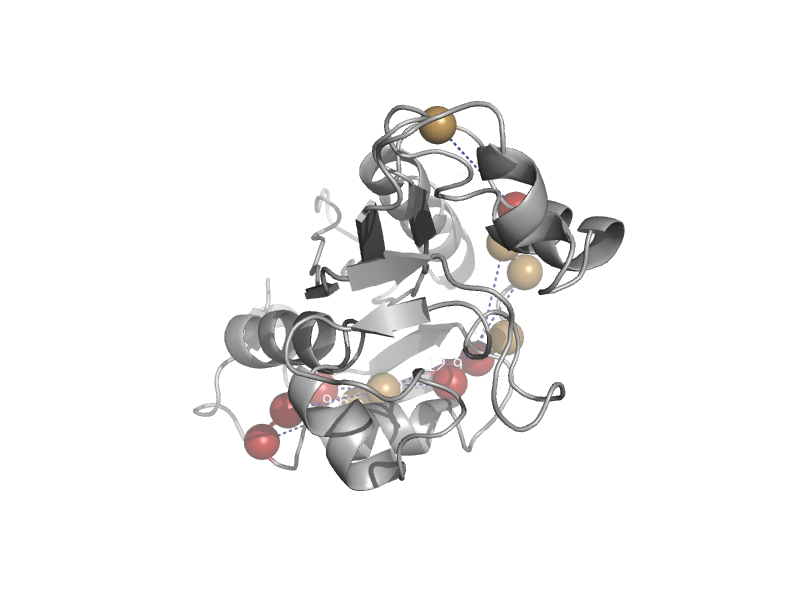 | view | |
| automated matches | automated matches | Escherichia coli K-12 [TaxId: 83333] | 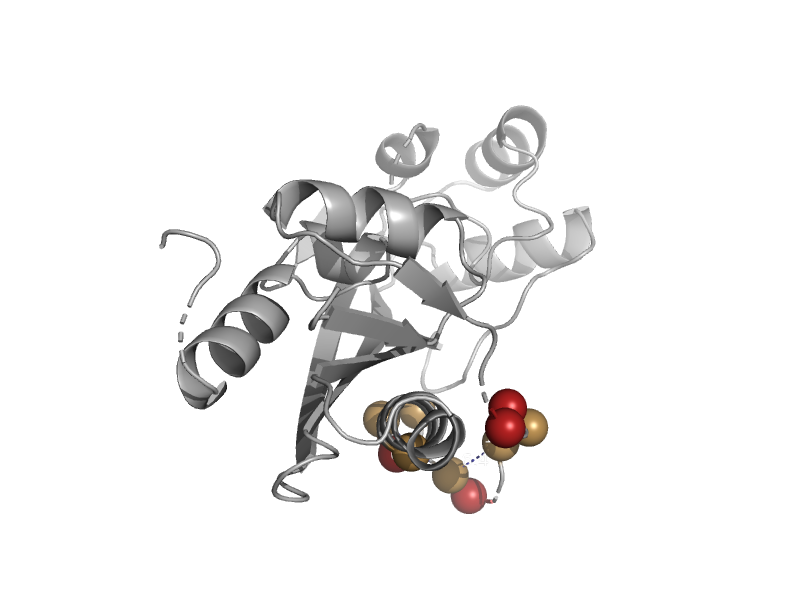 | view | |
| automated matches | automated matches | Burkholderia cenocepacia [TaxId: 216591] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | M | T | K | P | L | I | L | A | S | Q | S | P | R | R | K | E | L | L | D | L | L | Q | - | - | - | - | - | L | - | P | Y | S | I | I | V | S | - | - | - | E | V | E | E | K | L | N | R | N | F | S | P | E | E | N | V | Q | W | L | A | K | Q | K | A | K | A | V | A | D | L | H | P | - | - | - | - | - | H | A | I | V | I | G | A | D | T | M | V | C | L | D | - | - | - | G | E | C | L | G | - | - | - | - | - | K | P | Q | - | - | - | - | D | Q | E | E | A | A | S | M | L | R | R | L | S | G | - | - | - | - | - | R | S | H | S | V | I | T | A | V | S | I | Q | A | - | E | N | - | - | H | S | E | T | F | Y | D | K | T | E | V | A | F | W | S | L | S | E | E | E | I | W | T | Y | I | E | T | K | E | P | M | D | K | A | G | A | Y | G | I | Q | - | - | - | - | G | - | R | G | A | L | F | V | K | K | I | D | G | - | - | - | - | D | Y | Y | S | V | M | G | L | P | I | S | K | T | M | R | A | L | R | H | F | - | - | - | - | - |
| - | - | - | - | - | - | - | Q | K | V | V | L | A | T | G | N | V | G | K | V | R | E | L | A | S | L | L | D | - | - | F | G | L | - | D | I | V | A | Q | T | D | L | G | V | D | S | A | E | E | T | G | L | - | - | - | - | - | - | T | F | I | E | N | A | I | L | K | A | R | H | A | A | K | V | T | A | - | - | - | - | - | - | L | P | A | I | A | D | D | S | G | L | A | V | D | V | L | G | G | A | P | - | G | I | Y | S | A | R | Y | S | G | E | D | A | T | D | Q | K | N | L | Q | K | L | L | E | T | K | - | D | V | P | D | D | Q | R | Q | A | R | F | H | C | V | L | V | Y | L | R | H | A | E | D | P | T | P | L | V | C | H | G | S | W | P | G | V | I | T | R | - | - | - | - | E | P | A | G | T | - | - | - | - | G | G | F | G | Y | D | P | I | F | F | V | P | - | - | - | S | E | G | K | T | A | A | E | L | - | - | - | - | T | - | E | E | K | S | A | I | S | H | R | G | Q | A | L | - | L | L | L | D | A | L | R | N | - | G | - | - | - | - |
| - | - | - | - | - | - | - | H | Q | V | I | S | A | T | T | N | P | A | K | I | Q | A | I | L | Q | A | F | E | E | I | F | G | E | G | S | C | H | I | T | P | V | - | - | - | A | V | E | S | G | V | P | E | Q | P | F | G | S | E | E | T | R | A | G | A | R | N | R | V | D | N | A | R | R | L | H | P | - | - | - | - | - | Q | A | D | F | W | V | A | I | E | A | G | I | D | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | A | T | F | S | W | V | V | I | D | N | G | V | - | - | - | - | Q | R | G | E | A | R | S | A | T | L | P | L | P | - | - | - | A | V | I | L | D | R | V | R | Q | G | E | A | L | G | P | V | S | Q | Y | T | G | I | D | E | I | G | R | K | E | G | A | I | G | V | F | T | - | - | - | - | A | G | K | L | T | R | S | S | V | Y | Y | Q | A | V | I | L | A | L | S | P | F | H | N | A | - | - | - | - | - |
| - | - | - | - | - | - | - | M | K | I | F | F | I | T | S | N | P | G | K | V | R | E | V | A | N | F | L | G | T | - | F | G | I | - | E | I | V | Q | L | K | - | - | - | H | E | Y | P | E | I | Q | A | E | - | - | - | - | - | - | K | L | E | D | V | V | D | F | G | I | S | W | L | K | G | K | V | P | - | - | - | - | - | - | E | P | F | M | I | E | D | S | G | L | F | I | E | S | L | K | G | F | P | - | G | V | Y | S | S | Y | V | Y | R | - | - | - | - | - | T | I | G | L | E | G | I | L | K | L | M | E | G | A | - | - | E | D | R | R | A | Y | F | K | S | V | I | G | F | Y | I | D | G | - | - | - | K | A | Y | K | F | S | G | V | T | W | G | R | I | S | N | - | - | - | - | E | K | R | G | T | - | - | - | - | H | G | F | G | Y | D | P | I | F | I | P | E | - | - | - | G | S | E | K | T | F | A | E | M | - | - | - | - | T | I | E | E | K | N | A | L | S | H | R | G | K | A | L | K | A | F | F | E | W | L | K | V | N | L | K | Y | - | - |
| - | - | - | - | - | - | K | L | T | V | Y | L | A | T | T | N | P | H | K | V | - | E | I | K | M | I | A | P | - | - | W | - | M | - | E | I | L | P | S | P | - | - | - | - | - | I | V | V | E | D | G | E | - | - | - | - | - | - | T | F | L | E | N | S | V | K | K | A | V | V | Y | G | K | - | L | K | - | - | - | - | - | - | H | P | V | M | A | D | D | S | G | L | V | I | Y | S | L | G | G | F | P | - | G | V | M | S | A | - | F | M | - | E | E | H | S | Y | - | E | K | M | R | T | I | L | K | M | L | E | G | K | D | - | - | - | R | R | A | A | F | V | C | S | A | T | F | F | D | P | V | E | - | N | T | L | I | S | V | E | D | R | V | E | G | R | I | A | N | - | - | - | - | E | I | R | G | T | - | - | - | - | G | G | F | G | Y | D | P | F | F | I | P | D | - | - | - | G | Y | D | K | T | F | G | E | I | - | - | - | - | P | - | H | L | K | E | K | I | S | H | R | S | K | A | F | R | K | L | F | S | V | L | E | K | I | L | - | - | - | - |
| - | - | - | - | E | E | I | R | T | M | I | I | G | T | S | S | A | F | R | A | N | V | L | R | E | H | F | G | D | R | - | F | R | - | N | F | V | L | L | P | P | - | - | - | D | I | D | E | K | A | Y | R | A | A | D | P | F | E | L | T | E | S | I | A | R | A | K | M | K | A | V | L | E | K | A | R | Q | H | - | - | - | P | A | I | A | L | T | F | D | Q | V | V | V | K | G | - | - | - | D | E | V | R | E | - | - | - | - | - | K | P | L | - | - | - | - | S | T | E | Q | C | R | S | F | I | A | S | Y | S | G | - | - | - | - | - | G | G | V | R | T | V | A | T | Y | A | L | C | V | V | G | T | E | - | N | V | L | V | A | H | N | E | T | E | T | F | F | S | K | F | G | D | D | I | V | E | R | T | L | E | R | G | A | C | M | N | S | A | G | G | L | V | V | E | - | - | - | - | D | E | D | M | S | R | H | V | V | R | I | V | G | - | - | - | - | T | S | Y | G | V | R | G | M | E | P | A | V | V | E | K | L | L | S | Q | L | - | - | - | - | - |
| M | A | A | S | L | V | G | K | K | I | V | F | V | T | G | N | A | K | K | L | E | E | V | V | Q | I | L | G | D | K | F | P | C | - | T | L | V | A | Q | K | - | - | - | I | D | L | P | E | Y | Q | G | - | - | - | - | - | - | - | E | P | D | E | I | S | I | Q | K | C | Q | E | A | V | R | Q | V | Q | - | - | - | - | - | - | G | P | V | L | V | E | D | T | C | L | C | F | N | A | L | G | G | L | P | G | - | P | Y | I | K | W | F | L | E | - | - | - | - | - | K | L | K | P | E | G | L | H | Q | L | L | A | G | F | - | - | E | D | K | S | A | Y | A | L | C | T | F | A | L | S | T | G | D | P | S | Q | P | V | R | L | F | R | G | R | T | S | G | R | I | V | - | - | - | - | - | A | P | R | G | C | - | - | - | - | Q | D | F | G | W | D | P | C | F | Q | P | D | - | - | - | G | Y | E | Q | T | Y | A | E | M | - | - | - | - | P | K | A | E | K | N | A | V | S | H | R | F | R | A | L | L | E | L | Q | E | Y | F | G | S | - | L | A | A | - | - |
| - | - | - | - | - | - | - | P | K | L | I | L | A | S | T | S | P | W | R | R | A | L | L | E | K | L | Q | - | - | - | - | - | I | - | S | F | E | C | A | A | P | - | - | - | - | V | D | E | T | P | R | - | D | E | S | P | R | Q | L | V | L | R | L | A | Q | E | K | A | Q | S | L | A | S | R | Y | P | - | - | - | - | - | D | H | L | I | I | G | S | D | Q | V | C | V | L | D | - | - | - | G | E | I | T | G | - | - | - | - | - | K | P | L | - | - | - | - | T | E | E | N | A | R | L | Q | L | R | K | A | S | G | - | - | - | - | - | N | I | V | T | F | Y | T | G | L | A | L | F | N | S | A | N | G | - | H | L | Q | T | E | V | E | P | F | D | V | H | F | R | H | L | S | E | A | E | I | D | N | Y | V | R | K | E | H | P | L | H | C | A | G | S | F | K | S | - | - | - | - | - | G | - | F | G | I | T | L | F | E | R | L | E | G | R | - | - | - | D | P | N | T | L | V | G | L | P | L | I | A | L | C | Q | M | L | R | R | E | G | N | P | L | M |
| - | A | P | T | P | L | F | Q | T | L | Y | L | A | S | - | S | P | R | R | Q | E | L | L | Q | Q | I | G | - | - | - | - | - | V | - | - | F | E | L | L | L | P | R | P | D | E | D | A | A | L | A | L | P | G | E | A | A | D | A | Y | V | R | R | V | T | V | A | K | A | E | A | A | R | A | R | L | V | A | S | G | K | P | A | A | P | V | L | V | A | D | T | T | V | T | I | D | - | - | - | G | A | I | L | G | - | - | - | - | - | K | P | T | - | - | - | - | D | A | D | D | A | L | A | M | L | T | R | L | A | G | - | - | - | - | - | R | E | H | A | V | L | T | A | V | A | V | I | D | A | S | G | E | - | L | L | P | P | A | L | S | R | S | S | V | R | F | A | A | A | S | R | D | A | Y | V | R | Y | V | E | T | G | E | P | F | G | K | A | G | A | Y | A | I | Q | - | - | - | - | G | - | R | A | A | E | F | I | E | R | I | D | G | - | - | - | - | S | H | S | G | I | M | G | L | P | L | F | E | T | A | A | L | L | R | T | A | R | V | A | F | - |
