solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1dnpa2 ( 1 ) ttHLVWFrqdLrlhdNlALaaACrns-----sArVLALYIATprq
d1iqra2 ( 2 ) gpLLVWHrgdlrLhdHpALleAlar------g-pVVGLVVLDpnn
d1np7a2 ( 1 ) mkhvppTVLVWFrnDlrlhdHePLhrAlks------glaItAVYCYdprq
d1owla2 ( 3 ) aPILFWHrrDlrLsdNiGLaaAraq------saqLIGLfCLDpqi
d1u3da2 ( 13 ) cSIVWFrrDlrvedNpALaaAvra-------gpVIALFVwApee
d2wq7a1 ( 2 ) dsqRsTLVHWFrkGlrLhdNpALshIFtaAnaapgryFVRPIfIldpgi
d6ptza1 ( 1 ) mp--hrTIHLFRkGlrlhdNpTLlaALes---Se---tIyPVyvLDrrf
bbbb aaaaaaaa bbbbbbb
60 70 80 90 100
d1dnpa2 ( 41 ) wa----t-hnmsprqAelinaqLngLQiaLAekGIpLlfrevddfvaSve
d1iqra2 ( 40 ) lktt--------prrrAwfLenVraLReaYrargGALWVleg----lPwe
d1np7a2 ( 45 ) FaqthqgfaktgpwrSnflqqsVqnLaesLqkvgNkLLvTtg----lPeq
d1owla2 ( 42 ) lqs-----admaparVaylqgcLqeLqqrYqqagSrLLLLqg----dPqh
d1u3da2 ( 50 ) eg----h-yhpgrvsrwwlknslaqLdssLrslgTÇLItkrst---dSVa
d2wq7a1 ( 51 ) ld----wm-qvgAnrwrflQqtLedLdnqLrklnsrLFVVrg----kpae
d6ptza1 ( 42 ) la----samhigalrwHflLqsLedLhknLsrlgArLLViqg----eyes
aaaaaaaaaaaaaaaaaaaa bbbbb aaa
110 120 130 140 150
d1dnpa2 ( 86 ) iVkqVCaensVthLfYnyqyevnerardveVeraLr--nvvCeGfdDsvi
d1iqra2 ( 78 ) kVPeAArrlkAkaVYALtshtpyGryrDgrVreaLp---vpLhllpaphl
d1np7a2 ( 91 ) vIPqIAkqInAktIYyhrevtqeeldvErnLvkqLtilgieAkGywgstl
d1owla2 ( 83 ) lIpqLAqqLqAeAVyWnqdiepyGrdrDgqVaaaLktagiravqlwDqll
d1u3da2 ( 92 ) sLldvvksTgAsqIffNhLydplSlvrdhrAkdvLtaqgiaVrsfnAdll
d2wq7a1 ( 92 ) VFprIFksWrVeMLTFEtdiepySvtrdaaVqklAkaegVrVethcshti
d6ptza1 ( 84 ) vLrdHvqkwnItQVTLDaEmepfykemeanIrrLGaelgFeVlsrvghsl
aaaaaaaa bbbbb aaaaaaaaaaaaaaa bbbbb
160 170 180 190 200
d1dnpa2 ( 134 ) LppgaVmtg-nhemykvftpfknawlkrlregmpe-cvaapkvrssgsie
d1iqra2 ( 125 ) lppd-lp-----rayrvytpfsrlyr------gaapplpppeaLpkgp--
d1np7a2 ( 141 ) chpedlpfsiqd-lpdlftkfrkdiek--kkisirpcffapsqLlpsp--
d1owla2 ( 133 ) HspdqI-lsgsgnpysvygpfwknwqa----qpkptpvatPteLvdLs--
d1u3da2 ( 142 ) YepweVtde-lgrpfsmfaafwerClsM--pydpespllppkkIisgdvs
d2wq7a1 ( 142 ) ynpelViaknlgkapityqkflgiveql---k-vpkvlgvPek-----Lk
d6ptza1 ( 134 ) ydtkrildlnggspPltykrflhIlsqL---gdpevpvrnLta---edFq
aaaaaaaaa
210 220 230
d1dnpa2 ( 182 ) pspsitLnyprqsfdtahf
d1iqra2 ( 161 ) ---eeg----------eIpredpg
d1np7a2 ( 186 ) ------------nikLeltapppefFpqin--f
d1owla2 ( 176 ) ---peqltaIaplllseLptl-kqlgfdwdggf
d1u3da2 ( 189 ) kçvadpLvF
d2wq7a1 ( 183 ) nMptppkdeveqkdsaaYdcPtmkqLVkrpeel
d6ptza1 ( 178 ) rCmSpePg-----laerYrVPvpadLeippqsl
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| DNA photolyase | Cryptochrome/photolyase, N-terminal domain | Escherichia coli [TaxId: 562] | 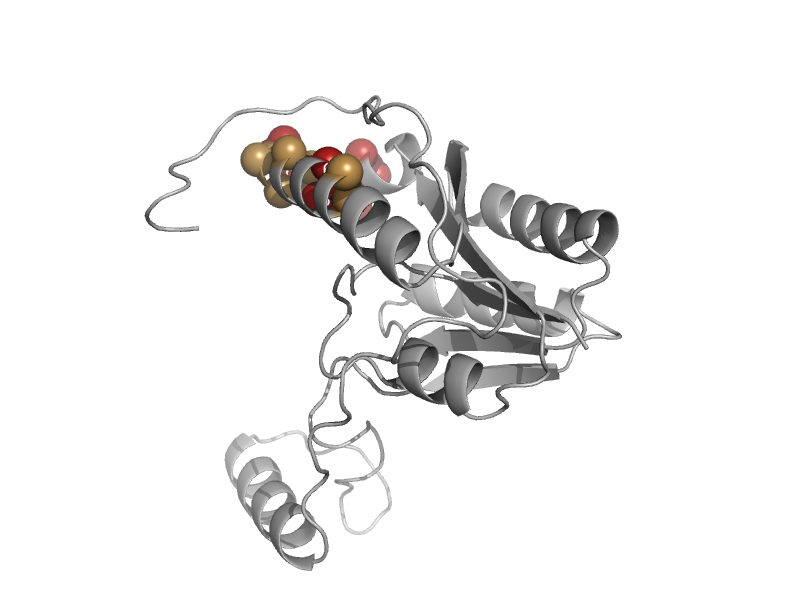 | view | |
| DNA photolyase | Cryptochrome/photolyase, N-terminal domain | Thermus thermophilus [TaxId: 274] | 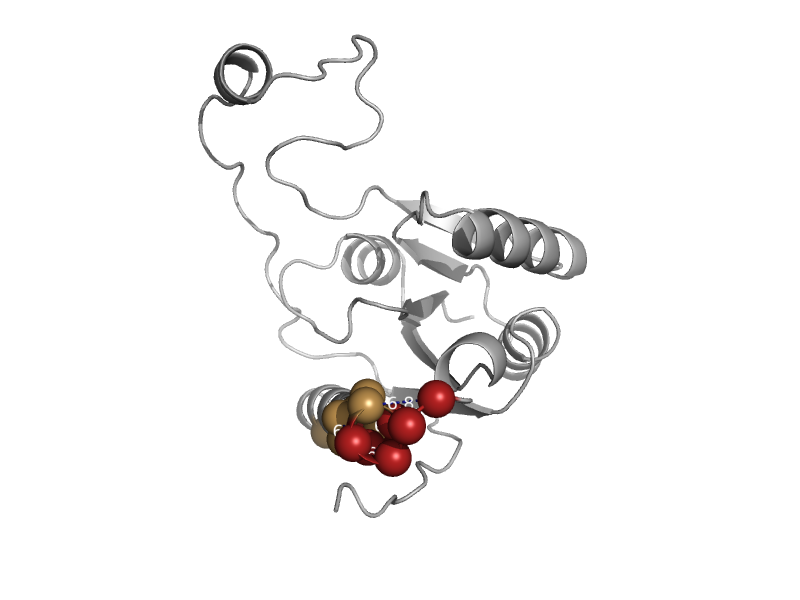 | view | |
| Cryptochrome | Cryptochrome/photolyase, N-terminal domain | Synechocystis sp. PCC 6803 [TaxId: 1148] | 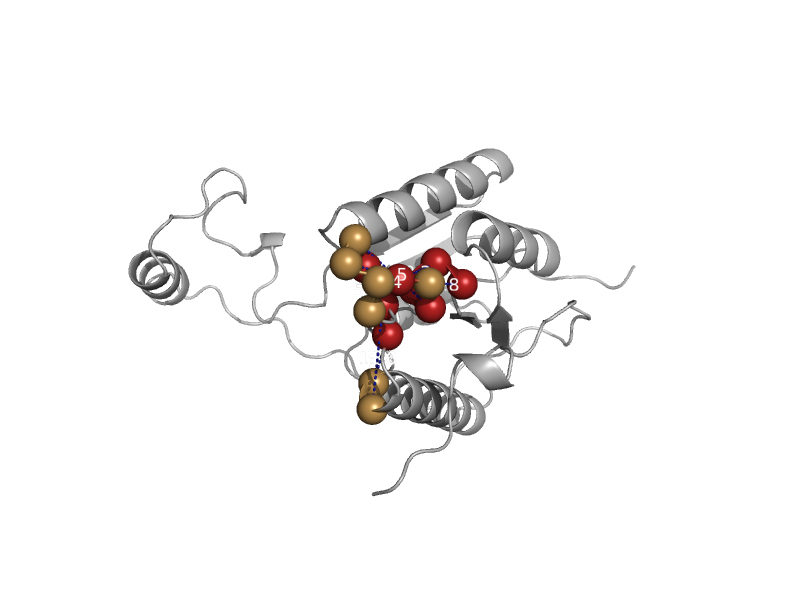 | view | |
| DNA photolyase | Cryptochrome/photolyase, N-terminal domain | Synechococcus elongatus [TaxId: 32046] | 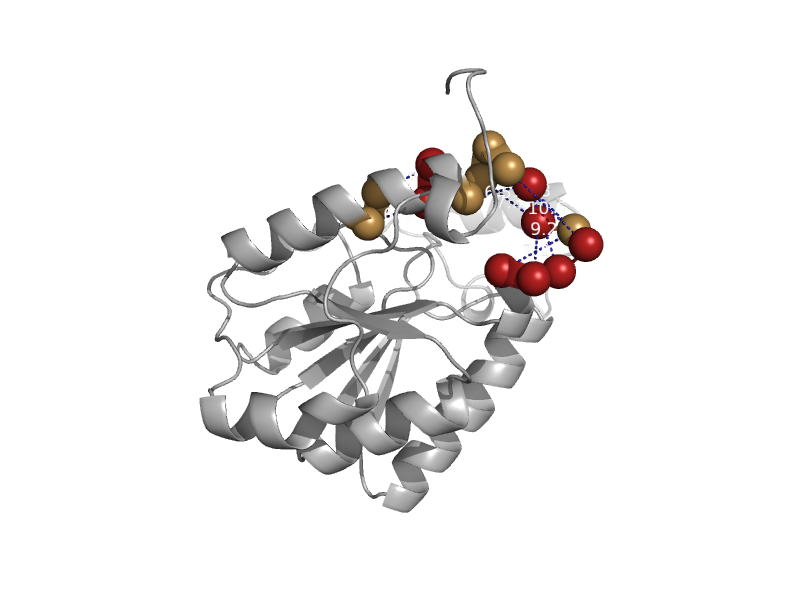 | view | |
| Cryptochrome | Cryptochrome/photolyase, N-terminal domain | Thale cress (Arabidopsis thaliana) [TaxId: 3702] |  | view | |
| automated matches | automated matches | Fruit fly (Drosophila melanogaster) [TaxId: 7227] |  | view | |
| automated matches | automated matches | Domestic pigeon (Columba livia) [TaxId: 8932] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | T | T | H | L | V | W | F | R | Q | D | L | R | L | H | D | N | L | A | L | A | A | A | C | R | N | S | - | - | - | - | - | S | A | R | V | L | A | L | Y | I | A | T | P | R | Q | W | A | - | - | - | - | T | - | H | N | M | S | P | R | Q | A | E | L | I | N | A | Q | L | N | G | L | Q | I | A | L | A | E | K | G | I | P | L | L | F | R | E | V | D | D | F | V | A | S | V | E | I | V | K | Q | V | C | A | E | N | S | V | T | H | L | F | Y | N | Y | Q | Y | E | V | N | E | R | A | R | D | V | E | V | E | R | A | L | R | - | - | N | V | V | C | E | G | F | D | D | S | V | I | L | P | P | G | A | V | M | T | G | - | N | H | E | M | Y | K | V | F | T | P | F | K | N | A | W | L | K | R | L | R | E | G | M | P | E | - | C | V | A | A | P | K | V | R | S | S | G | S | I | E | P | S | P | S | I | T | L | N | Y | P | R | Q | S | F | D | T | A | H | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | G | P | L | L | V | W | H | R | G | D | L | R | L | H | D | H | P | A | L | L | E | A | L | A | R | - | - | - | - | - | - | G | - | P | V | V | G | L | V | V | L | D | P | N | N | L | K | T | T | - | - | - | - | - | - | - | - | P | R | R | R | A | W | F | L | E | N | V | R | A | L | R | E | A | Y | R | A | R | G | G | A | L | W | V | L | E | G | - | - | - | - | L | P | W | E | K | V | P | E | A | A | R | R | L | K | A | K | A | V | Y | A | L | T | S | H | T | P | Y | G | R | Y | R | D | G | R | V | R | E | A | L | P | - | - | - | V | P | L | H | L | L | P | A | P | H | L | L | P | P | D | - | L | P | - | - | - | - | - | R | A | Y | R | V | Y | T | P | F | S | R | L | Y | R | - | - | - | - | - | - | G | A | A | P | P | L | P | P | P | E | A | L | P | K | G | P | - | - | - | - | - | E | E | G | - | - | - | - | - | - | - | - | - | - | E | I | P | R | E | D | P | G | - | - | - | - | - | - | - | - | - |
| M | K | H | V | P | P | T | V | L | V | W | F | R | N | D | L | R | L | H | D | H | E | P | L | H | R | A | L | K | S | - | - | - | - | - | - | G | L | A | I | T | A | V | Y | C | Y | D | P | R | Q | F | A | Q | T | H | Q | G | F | A | K | T | G | P | W | R | S | N | F | L | Q | Q | S | V | Q | N | L | A | E | S | L | Q | K | V | G | N | K | L | L | V | T | T | G | - | - | - | - | L | P | E | Q | V | I | P | Q | I | A | K | Q | I | N | A | K | T | I | Y | Y | H | R | E | V | T | Q | E | E | L | D | V | E | R | N | L | V | K | Q | L | T | I | L | G | I | E | A | K | G | Y | W | G | S | T | L | C | H | P | E | D | L | P | F | S | I | Q | D | - | L | P | D | L | F | T | K | F | R | K | D | I | E | K | - | - | K | K | I | S | I | R | P | C | F | F | A | P | S | Q | L | L | P | S | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | I | K | L | E | L | T | A | P | P | P | E | F | F | P | Q | I | N | - | - | F |
| - | - | - | - | - | A | P | I | L | F | W | H | R | R | D | L | R | L | S | D | N | I | G | L | A | A | A | R | A | Q | - | - | - | - | - | - | S | A | Q | L | I | G | L | F | C | L | D | P | Q | I | L | Q | S | - | - | - | - | - | A | D | M | A | P | A | R | V | A | Y | L | Q | G | C | L | Q | E | L | Q | Q | R | Y | Q | Q | A | G | S | R | L | L | L | L | Q | G | - | - | - | - | D | P | Q | H | L | I | P | Q | L | A | Q | Q | L | Q | A | E | A | V | Y | W | N | Q | D | I | E | P | Y | G | R | D | R | D | G | Q | V | A | A | A | L | K | T | A | G | I | R | A | V | Q | L | W | D | Q | L | L | H | S | P | D | Q | I | - | L | S | G | S | G | N | P | Y | S | V | Y | G | P | F | W | K | N | W | Q | A | - | - | - | - | Q | P | K | P | T | P | V | A | T | P | T | E | L | V | D | L | S | - | - | - | - | - | P | E | Q | L | T | A | I | A | P | L | L | L | S | E | L | P | T | L | - | K | Q | L | G | F | D | W | D | G | G | F |
| - | - | - | - | - | - | C | S | I | V | W | F | R | R | D | L | R | V | E | D | N | P | A | L | A | A | A | V | R | A | - | - | - | - | - | - | - | G | P | V | I | A | L | F | V | W | A | P | E | E | E | G | - | - | - | - | H | - | Y | H | P | G | R | V | S | R | W | W | L | K | N | S | L | A | Q | L | D | S | S | L | R | S | L | G | T | C | L | I | T | K | R | S | T | - | - | - | D | S | V | A | S | L | L | D | V | V | K | S | T | G | A | S | Q | I | F | F | N | H | L | Y | D | P | L | S | L | V | R | D | H | R | A | K | D | V | L | T | A | Q | G | I | A | V | R | S | F | N | A | D | L | L | Y | E | P | W | E | V | T | D | E | - | L | G | R | P | F | S | M | F | A | A | F | W | E | R | C | L | S | M | - | - | P | Y | D | P | E | S | P | L | L | P | P | K | K | I | I | S | G | D | V | S | K | C | V | A | D | P | L | V | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | D | S | Q | R | S | T | L | V | H | W | F | R | K | G | L | R | L | H | D | N | P | A | L | S | H | I | F | T | A | A | N | A | A | P | G | R | Y | F | V | R | P | I | F | I | L | D | P | G | I | L | D | - | - | - | - | W | M | - | Q | V | G | A | N | R | W | R | F | L | Q | Q | T | L | E | D | L | D | N | Q | L | R | K | L | N | S | R | L | F | V | V | R | G | - | - | - | - | K | P | A | E | V | F | P | R | I | F | K | S | W | R | V | E | M | L | T | F | E | T | D | I | E | P | Y | S | V | T | R | D | A | A | V | Q | K | L | A | K | A | E | G | V | R | V | E | T | H | C | S | H | T | I | Y | N | P | E | L | V | I | A | K | N | L | G | K | A | P | I | T | Y | Q | K | F | L | G | I | V | E | Q | L | - | - | - | K | - | V | P | K | V | L | G | V | P | E | K | - | - | - | - | - | L | K | N | M | P | T | P | P | K | D | E | V | E | Q | K | D | S | A | A | Y | D | C | P | T | M | K | Q | L | V | K | R | P | E | E | L |
| - | M | P | - | - | H | R | T | I | H | L | F | R | K | G | L | R | L | H | D | N | P | T | L | L | A | A | L | E | S | - | - | - | S | E | - | - | - | T | I | Y | P | V | Y | V | L | D | R | R | F | L | A | - | - | - | - | S | A | M | H | I | G | A | L | R | W | H | F | L | L | Q | S | L | E | D | L | H | K | N | L | S | R | L | G | A | R | L | L | V | I | Q | G | - | - | - | - | E | Y | E | S | V | L | R | D | H | V | Q | K | W | N | I | T | Q | V | T | L | D | A | E | M | E | P | F | Y | K | E | M | E | A | N | I | R | R | L | G | A | E | L | G | F | E | V | L | S | R | V | G | H | S | L | Y | D | T | K | R | I | L | D | L | N | G | G | S | P | P | L | T | Y | K | R | F | L | H | I | L | S | Q | L | - | - | - | G | D | P | E | V | P | V | R | N | L | T | A | - | - | - | E | D | F | Q | R | C | M | S | P | E | P | G | - | - | - | - | - | L | A | E | R | Y | R | V | P | V | P | A | D | L | E | I | P | P | Q | S | L |
