solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1f8ya- ( 2 ) pkktIYFGAgw-f-----tdrqnkAykeAmeaLkeNptIdlenSyvPld
d1isia- ( 2 ) wraeGTsa----hLrdiFlgrÇaeYralLspeq--r-n--kdçta
d1s2la- ( 3 ) aptGkIYLGSpf-s------daq-erAa--AklLa-NpsIa--hvfFpfd
d1t1ja1 ( 1 ) mrkIFLACPyshadaevveqrfraCnevaatIvragh----vvfsqVs
d2f62a2 ( 10 ) rkIYIAGpavf-----npdGasyYnkVrelLkke-----nVplIptd
d3f6ya- ( 45 ) fwrqtwsGpGTtk----rFpetVlarÇvkyteih-peMr---h--Vdçqs
d3i9la- ( 1 ) ivPtr----eLenvFlgrÇkdYeitryldiLprvr--sdçsa
d6evsa- ( 2 ) akIYLASPf-f-----neeQlkhVskAeqVLrdlgh----tVfSpre
bb aaaaaaaaaaaaa
60 70 80 90 100
d1f8ya- ( 45 ) nqykgirvdehpeylh-dkvwatatynndlnGIktndIMLGVYIPd---e
d1isia- ( 38 ) IWeaFkvAL-dkdPçsVlpsdYdlFItlSrHsIprdkSLFwenshllVns
d1s2la- ( 48 ) gftDpdepigg---ir-smvwrdatyqndltgIsnAtCGVFLydmd---q
d1t1ja1 ( 45 ) mSHpinlclaeld----raaigrlwapvDafyMdhleeLIVLdlp--gwr
d2f62a2 ( 49 ) neat----------------ealdirqkniq-IkdCdaVIAdLspfrghe
d3f6ya- ( 85 ) VwdaFkgAFiskhPçdIteedYqpLMklGtqtvpçnkIlLwSrik---ft
d3i9la- ( 37 ) LwkdFfkAFsfknpçdldlgsYkdFFtsAqQqLpknkVMfwsgVydeAhd
d6evsa- ( 39 ) nqlp------evefg--sfewrtfvfkndlehIkwAdITFGIigdn---y
aaaaaaa bbbb
110 120 130 140 150
d1f8ya- ( 91 ) ed------------------------------------------------
d1isia- ( 87 ) FAdntrrfmPlsdVLYGrVAdflsWÇrqkadsgldyqsÇPts-edçenNP
d1s2la- ( 94 ) ld------------------------------------------------
d1t1ja1 ( 89 ) ds------------------------------------------------
d2f62a2 ( 83 ) Pd------------------------------------------------
d3f6ya- ( 137 ) qvq--rdmftLedtLLGyLAddLtWÇGefdtskInyqsÇPdwrkdçsnnP
d3i9la- ( 87 ) yAntGrkyITleDTLPGyMLnslvwçGqranpGfnekvÇpdf-ktçpvqA
d6evsa- ( 78 ) dd------------------------------------------------
160 170 180 190 200
d1f8ya- ( 93 ) --vgLgmeLgyAlsqg-kyVLLVIPdeDygk-------------------
d1isia- ( 136 ) vdSFwkrASiqYSkdSsGvIhVMLnGSep-tGAYpikgfFAdyeIpnLqk
d1s2la- ( 96 ) --dgSafeIgfMramh-kpVILVPftp-----------------------
d1t1ja1 ( 91 ) --agIrrEmefFeagg-qrvSl---wseVeheF
d2f62a2 ( 85 ) --cgTafevgcAaaln-k-VLtfTsdrrnreKygsgvdk-----------
d3f6ya- ( 185 ) vsVFwkTVSrrFAeaAçdvVhVmLdGsrs--kIFdkdstFGsVEVhnLqp
d3i9la- ( 136 ) reSFwGmAsssYAhsAeGeVTYMVdGSnpkvpAYrpdsfFGkyGLpnLt-
d6evsa- ( 80 ) --tgTaweLgaSyilg-kpVMLFSp---tge-------------------
aaaaaaaaaaaa bbbb
210 220 230 240 250
d1f8ya- ( 121 ) ------------------------pInlmswgvS----dnVi---kmsqL
d1isia- ( 185 ) ekItrIeIWVMheIggpnvesçgegsMkvLekrLkdmgFqYsçindyrpV
d1s2la- ( 122 ) ------------------------eMnlmiAqgV----tTiIdGnefekL
d1t1ja1
d2f62a2 ( 122 ) --------------dnlrvegfglPfnl-lydgv-----evfd--sFesA
d3f6ya- ( 233 ) ekVqtLeAwVihg-g----dlçqdptIkeLesiIskrnIqFsÇkniy
d3i9la- ( 185 ) nkVtrVkViVLHrLgekiiekçgagsLldLeklVkakhFaFdÇvenpraV
d6evsa- ( 105 ) ------------------------iinlmitdsL----hAyFe--dwndV
aaaa bb
260
d1f8ya- ( 140 ) kdfnFnkprfdfYe-gavy
d1isia- ( 235 ) kllqçvdhs--thpdÇalk
d1s2la- ( 149 ) adynFn-cpsnpVrgygiy
d1t1ja1
d2f62a2 ( 151 ) FkyFlanfps
d3f6ya-
d3i9la- ( 235 ) lfllçsdnp--nareÇrla
d6evsa- ( 125 ) enYdfatlpikpyl
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Nucleoside 2-deoxyribosyltransferase | N-deoxyribosyltransferase | Lactobacillus leichmannii [TaxId: 28039] | 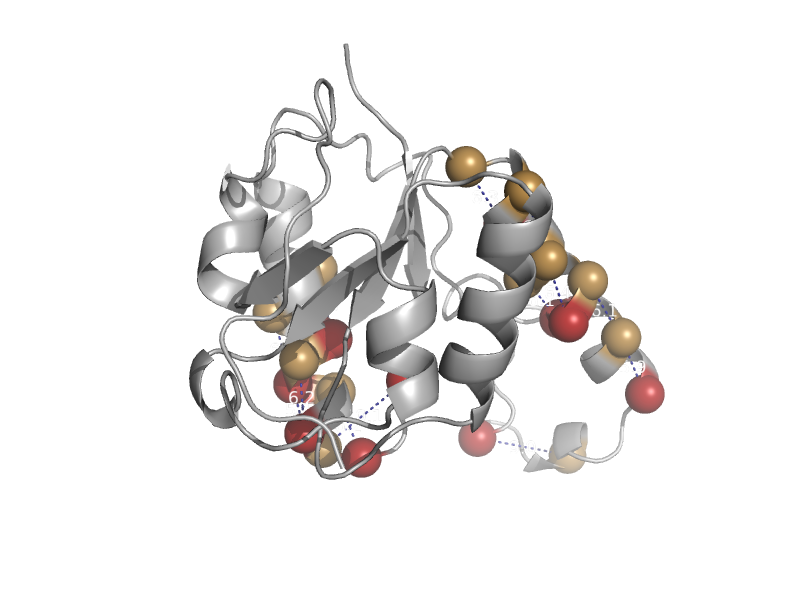 | view | |
| Bone marror stromal cell antigen 1, BST-1/CD157 (ADP ribosyl cyclase-2) | ADP ribosyl cyclase-like | Human (Homo sapiens) [TaxId: 9606] | 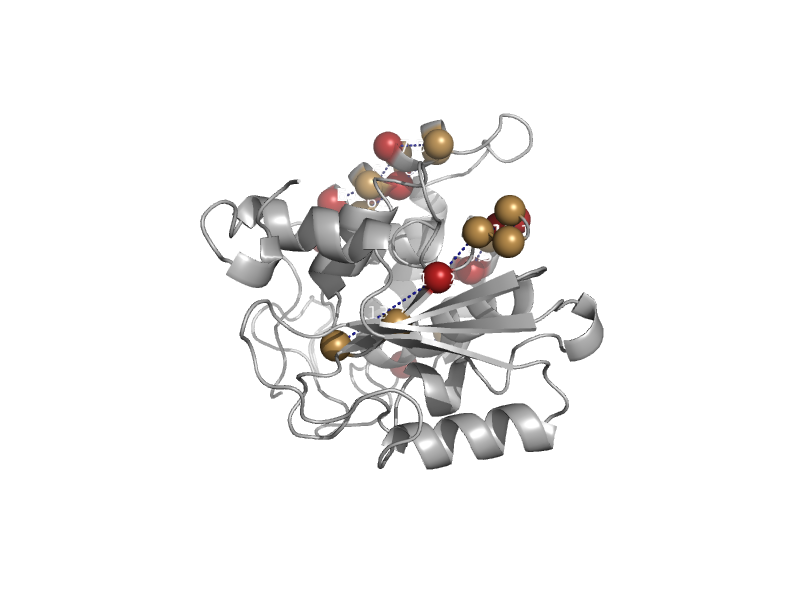 | view | |
| Purine transdeoxyribosylase | N-deoxyribosyltransferase | Lactobacillus helveticus [TaxId: 1587] | 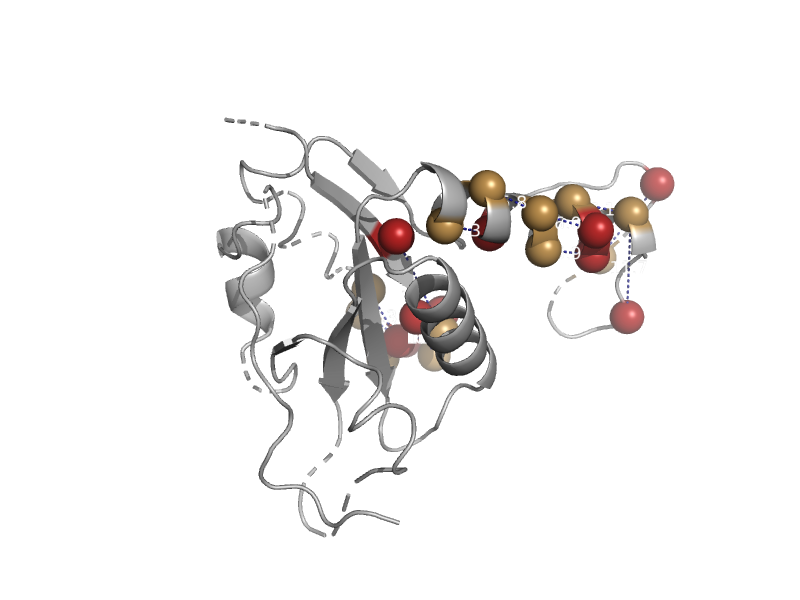 | view | |
| Hypothetical protein PA1492 | Hypothetical protein PA1492 | Pseudomonas aeruginosa [TaxId: 287] | 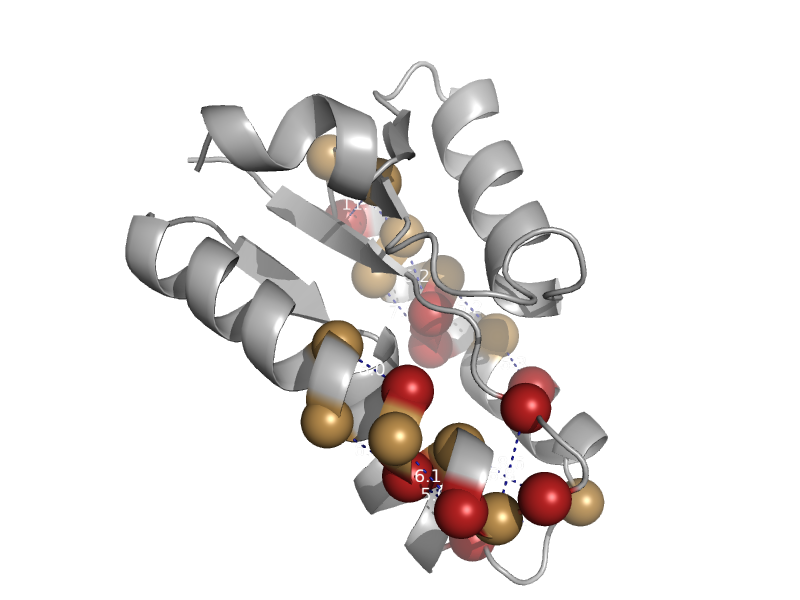 | view | |
| Nucleoside 2-deoxyribosyltransferase | N-deoxyribosyltransferase | Trypanosome (Trypanosoma brucei) [TaxId: 5691] | 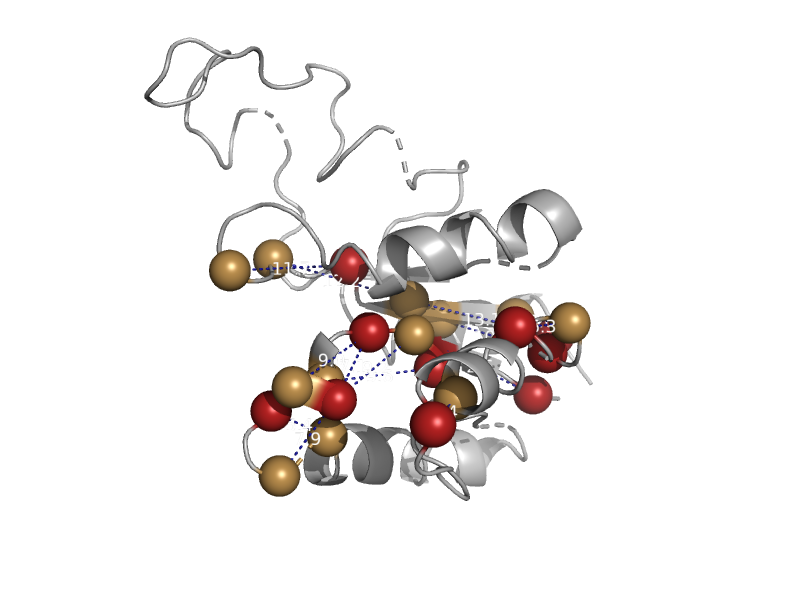 | view | |
| ADP ribosyl cyclase | ADP ribosyl cyclase-like | Human (Homo sapiens) [TaxId: 9606] | 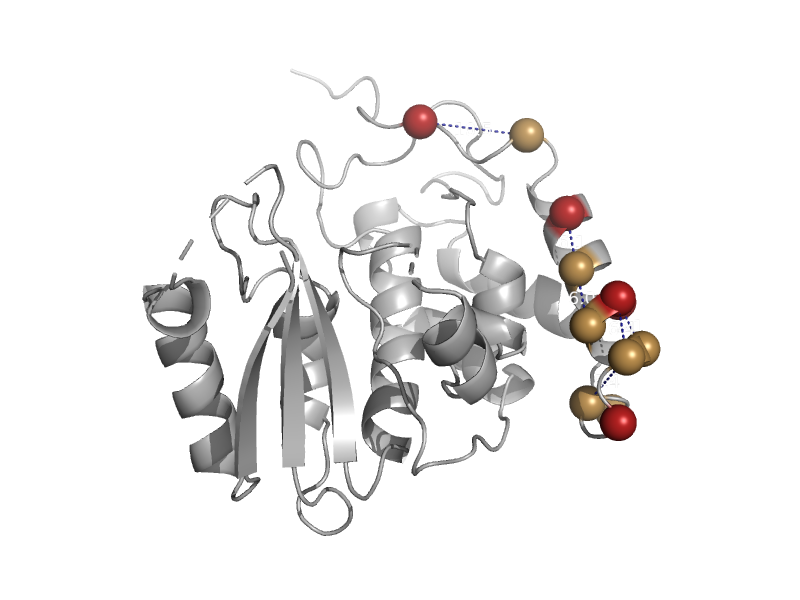 | view | |
| automated matches | ADP ribosyl cyclase-like | California sea hare (Aplysia californica) [TaxId: 6500] | 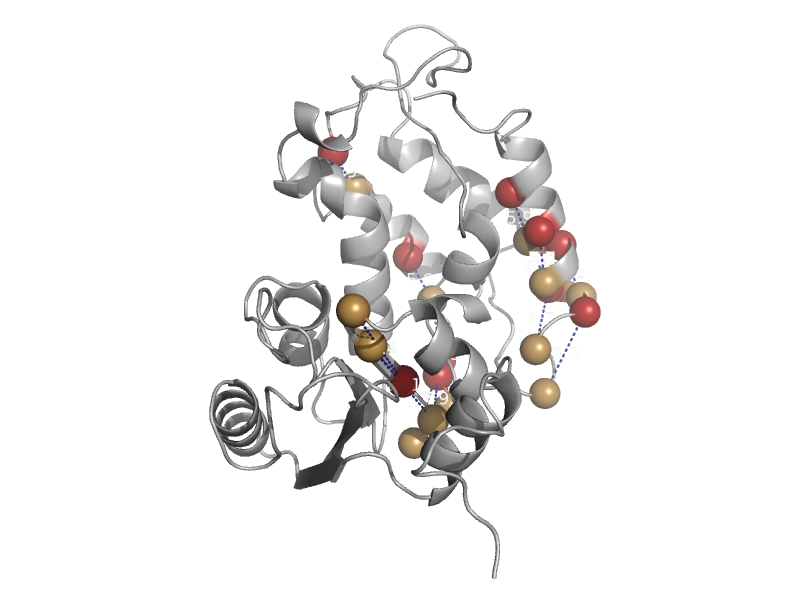 | view | |
| automated matches | automated matches | Bacillus psychrosaccharolyticus [TaxId: 1407] | 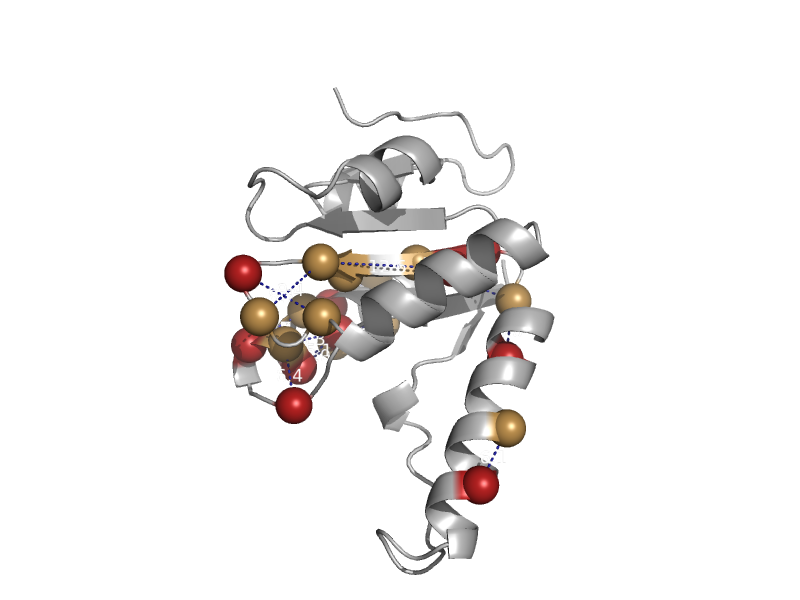 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | P | K | K | T | I | Y | F | G | A | G | W | - | F | - | - | - | - | - | T | D | R | Q | N | K | A | Y | K | E | A | M | E | A | L | K | E | N | P | T | I | D | L | E | N | S | Y | V | P | L | D | N | Q | Y | K | G | I | R | V | D | E | H | P | E | Y | L | H | - | D | K | V | W | A | T | A | T | Y | N | N | D | L | N | G | I | K | T | N | D | I | M | L | G | V | Y | I | P | D | - | - | - | E | E | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V | G | L | G | M | E | L | G | Y | A | L | S | Q | G | - | K | Y | V | L | L | V | I | P | D | E | D | Y | G | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | I | N | L | M | S | W | G | V | S | - | - | - | - | D | N | V | I | - | - | - | K | M | S | Q | L | K | D | F | N | F | N | K | P | R | F | D | F | Y | E | - | G | A | V | Y |
| - | - | - | - | - | W | R | A | E | G | T | S | A | - | - | - | - | H | L | R | D | I | F | L | G | R | C | A | E | Y | R | A | L | L | S | P | E | Q | - | - | R | - | N | - | - | K | D | C | T | A | I | W | E | A | F | K | V | A | L | - | D | K | D | P | C | S | V | L | P | S | D | Y | D | L | F | I | T | L | S | R | H | S | I | P | R | D | K | S | L | F | W | E | N | S | H | L | L | V | N | S | F | A | D | N | T | R | R | F | M | P | L | S | D | V | L | Y | G | R | V | A | D | F | L | S | W | C | R | Q | K | A | D | S | G | L | D | Y | Q | S | C | P | T | S | - | E | D | C | E | N | N | P | V | D | S | F | W | K | R | A | S | I | Q | Y | S | K | D | S | S | G | V | I | H | V | M | L | N | G | S | E | P | - | T | G | A | Y | P | I | K | G | F | F | A | D | Y | E | I | P | N | L | Q | K | E | K | I | T | R | I | E | I | W | V | M | H | E | I | G | G | P | N | V | E | S | C | G | E | G | S | M | K | V | L | E | K | R | L | K | D | M | G | F | Q | Y | S | C | I | N | D | Y | R | P | V | K | L | L | Q | C | V | D | H | S | - | - | T | H | P | D | C | A | L | K |
| A | P | T | G | K | I | Y | L | G | S | P | F | - | S | - | - | - | - | - | - | D | A | Q | - | E | R | A | A | - | - | A | K | L | L | A | - | N | P | S | I | A | - | - | H | V | F | F | P | F | D | G | F | T | D | P | D | E | P | I | G | G | - | - | - | I | R | - | S | M | V | W | R | D | A | T | Y | Q | N | D | L | T | G | I | S | N | A | T | C | G | V | F | L | Y | D | M | D | - | - | - | Q | L | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | G | S | A | F | E | I | G | F | M | R | A | M | H | - | K | P | V | I | L | V | P | F | T | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | M | N | L | M | I | A | Q | G | V | - | - | - | - | T | T | I | I | D | G | N | E | F | E | K | L | A | D | Y | N | F | N | - | C | P | S | N | P | V | R | G | Y | G | I | Y |
| - | - | M | R | K | I | F | L | A | C | P | Y | S | H | A | D | A | E | V | V | E | Q | R | F | R | A | C | N | E | V | A | A | T | I | V | R | A | G | H | - | - | - | - | V | V | F | S | Q | V | S | M | S | H | P | I | N | L | C | L | A | E | L | D | - | - | - | - | R | A | A | I | G | R | L | W | A | P | V | D | A | F | Y | M | D | H | L | E | E | L | I | V | L | D | L | P | - | - | G | W | R | D | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | G | I | R | R | E | M | E | F | F | E | A | G | G | - | Q | R | V | S | L | - | - | - | W | S | E | V | E | H | E | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | R | K | I | Y | I | A | G | P | A | V | F | - | - | - | - | - | N | P | D | G | A | S | Y | Y | N | K | V | R | E | L | L | K | K | E | - | - | - | - | - | N | V | P | L | I | P | T | D | N | E | A | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | A | L | D | I | R | Q | K | N | I | Q | - | I | K | D | C | D | A | V | I | A | D | L | S | P | F | R | G | H | E | P | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | G | T | A | F | E | V | G | C | A | A | A | L | N | - | K | - | V | L | T | F | T | S | D | R | R | N | R | E | K | Y | G | S | G | V | D | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | N | L | R | V | E | G | F | G | L | P | F | N | L | - | L | Y | D | G | V | - | - | - | - | - | E | V | F | D | - | - | S | F | E | S | A | F | K | Y | F | L | A | N | F | P | S | - | - | - | - | - | - | - | - | - |
| F | W | R | Q | T | W | S | G | P | G | T | T | K | - | - | - | - | R | F | P | E | T | V | L | A | R | C | V | K | Y | T | E | I | H | - | P | E | M | R | - | - | - | H | - | - | V | D | C | Q | S | V | W | D | A | F | K | G | A | F | I | S | K | H | P | C | D | I | T | E | E | D | Y | Q | P | L | M | K | L | G | T | Q | T | V | P | C | N | K | I | L | L | W | S | R | I | K | - | - | - | F | T | Q | V | Q | - | - | R | D | M | F | T | L | E | D | T | L | L | G | Y | L | A | D | D | L | T | W | C | G | E | F | D | T | S | K | I | N | Y | Q | S | C | P | D | W | R | K | D | C | S | N | N | P | V | S | V | F | W | K | T | V | S | R | R | F | A | E | A | A | C | D | V | V | H | V | M | L | D | G | S | R | S | - | - | K | I | F | D | K | D | S | T | F | G | S | V | E | V | H | N | L | Q | P | E | K | V | Q | T | L | E | A | W | V | I | H | G | - | G | - | - | - | - | D | L | C | Q | D | P | T | I | K | E | L | E | S | I | I | S | K | R | N | I | Q | F | S | C | K | N | I | Y | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | I | V | P | T | R | - | - | - | - | E | L | E | N | V | F | L | G | R | C | K | D | Y | E | I | T | R | Y | L | D | I | L | P | R | V | R | - | - | S | D | C | S | A | L | W | K | D | F | F | K | A | F | S | F | K | N | P | C | D | L | D | L | G | S | Y | K | D | F | F | T | S | A | Q | Q | Q | L | P | K | N | K | V | M | F | W | S | G | V | Y | D | E | A | H | D | Y | A | N | T | G | R | K | Y | I | T | L | E | D | T | L | P | G | Y | M | L | N | S | L | V | W | C | G | Q | R | A | N | P | G | F | N | E | K | V | C | P | D | F | - | K | T | C | P | V | Q | A | R | E | S | F | W | G | M | A | S | S | S | Y | A | H | S | A | E | G | E | V | T | Y | M | V | D | G | S | N | P | K | V | P | A | Y | R | P | D | S | F | F | G | K | Y | G | L | P | N | L | T | - | N | K | V | T | R | V | K | V | I | V | L | H | R | L | G | E | K | I | I | E | K | C | G | A | G | S | L | L | D | L | E | K | L | V | K | A | K | H | F | A | F | D | C | V | E | N | P | R | A | V | L | F | L | L | C | S | D | N | P | - | - | N | A | R | E | C | R | L | A |
| - | - | - | A | K | I | Y | L | A | S | P | F | - | F | - | - | - | - | - | N | E | E | Q | L | K | H | V | S | K | A | E | Q | V | L | R | D | L | G | H | - | - | - | - | T | V | F | S | P | R | E | N | Q | L | P | - | - | - | - | - | - | E | V | E | F | G | - | - | S | F | E | W | R | T | F | V | F | K | N | D | L | E | H | I | K | W | A | D | I | T | F | G | I | I | G | D | N | - | - | - | Y | D | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | G | T | A | W | E | L | G | A | S | Y | I | L | G | - | K | P | V | M | L | F | S | P | - | - | - | T | G | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | I | I | N | L | M | I | T | D | S | L | - | - | - | - | H | A | Y | F | E | - | - | D | W | N | D | V | E | N | Y | D | F | A | T | L | P | I | K | P | Y | L | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1f8ya_ | A | Q9R5V5 | GO:0009117 |
| d1f8ya_ | A | Q9R5V5 | GO:0016740 |
| d1f8ya_ | A | Q9R5V5 | GO:0043173 |
| d1f8ya_ | A | Q9R5V5 | GO:0050144 |
| d1isia_ | A | Q10588 | GO:0001931 |
| d1isia_ | A | Q10588 | GO:0001952 |
| d1isia_ | A | Q10588 | GO:0002691 |
| d1isia_ | A | Q10588 | GO:0003953 |
| d1isia_ | A | Q10588 | GO:0005576 |
| d1isia_ | A | Q10588 | GO:0005886 |
| d1isia_ | A | Q10588 | GO:0006959 |
| d1isia_ | A | Q10588 | GO:0007165 |
| d1isia_ | A | Q10588 | GO:0008284 |
| d1isia_ | A | Q10588 | GO:0016020 |
| d1isia_ | A | Q10588 | GO:0016740 |
| d1isia_ | A | Q10588 | GO:0016787 |
| d1isia_ | A | Q10588 | GO:0016798 |
| d1isia_ | A | Q10588 | GO:0016849 |
| d1isia_ | A | Q10588 | GO:0030890 |
| d1isia_ | A | Q10588 | GO:0032956 |
| d1isia_ | A | Q10588 | GO:0035579 |
| d1isia_ | A | Q10588 | GO:0050727 |
| d1isia_ | A | Q10588 | GO:0050730 |
| d1isia_ | A | Q10588 | GO:0050848 |
| d1isia_ | A | Q10588 | GO:0061809 |
| d1isia_ | A | Q10588 | GO:0070062 |
| d1isia_ | A | Q10588 | GO:0090022 |
| d1isia_ | A | Q10588 | GO:0090322 |
| d1isia_ | A | Q10588 | GO:0098552 |
| d1isia_ | A | Q10588 | GO:2001044 |
| d1s2la_ | A | Q8RLY5 | GO:0016757 |
| d1s2la_ | A | Q8RLY5 | GO:0050144 |
