solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1muga- ( 1 )
d1oe4a- ( 36 ) esPAdsFLkvEleLnlkLsnlvFqdpVqyvYnPLvYAwa
d1vk2a- ( 2 ) ytreeleivserVkct------------acpLhl
d1wywa- ( 117 ) ae
d2c53a- ( 18 ) dWttfrvFlIddaWrpLMepElanplTahLlaeYnrrcq--teeVlPpre
d2hxma2 ( 85 ) fgesWkkhLsgefgkpyfikLmgfvaeerk--hytVyPpph
d2q07a2 ( 244 )
d4wpka1 ( 1 ) MarplselVergWaaaLepva--dqVahMgqfLraeia-agrrylPags
60 70 80 90 100
d1muga- ( 1 ) mVedILapgLrVVFCgiNPgls-sagtgfpfAhp------------
d1oe4a- ( 75 ) PHenYVqtYCkskKeVLFLGmNPgpfGMAQTGVPfGEvnhVrdWLqIegp
d1vk2a- ( 26 ) nrtnVVvGegnldTrIVFVGeGPgee-ed-tgrPFvgr------------
d1wywa- ( 119 ) lltktLpdiLtfnLdIVIIgiNPglm-aAykghHyPgp------------
d2c53a- ( 67 ) dVFSWTryCtPdeVrVVIIgqnPyhhpgqAhgLAfSVa------------
d2hxma2 ( 124 ) qvftWTqmcdIkdVkVVILgqdPyhgpnqAHgLCfSVqr-----------
d2q07a2 ( 244 )
d4wpka1 ( 48 ) nVLrAFt-fpfdnVrVLIVGqDPyptpghAvgLSfSVap-----------
bbbbb
110 120 130 140 150
d1muga- ( 34 ) ------------------------anrFWkVIyqAg-FTdrqLk------
d1oe4a- ( 125 ) VskpevehpkrrIrGfeCpqsevsGarFWsLFkslC-g------------
d1vk2a- ( 63 ) ------------------------aGlLteLLresg------ir------
d1wywa- ( 156 ) ------------------------gnhFWkCLfmSg-LSevqln------
d2c53a- ( 106 ) ----------------nv-ppppsLrnVlaAVknCyp-------------
d2hxma2 ( 163 ) ----------------pv-ppppsLenIYkELstDie------------d
d2q07a2 ( 244 ) ryfFerAlecYkpfsdTVLLLpctar
d4wpka1 ( 86 ) ----------------dvrpwprsLanIFdEYtaDlg-------------
aaaaaaa
160 170 180 190 200
d1muga- ( 53 ) -----pqeAqhLl-d---yrCGVTkLvdrpt-v------qaneVs----k
d1oe4a- ( 162 ) -------qPetFFk-----hCFVhNHCPLIFMnhsGknltPtdLpkaqrd
d1vk2a- ( 78 ) --------red---------VyICNVVKcrP-p------nnrtPt----p
d1wywa- ( 175 ) -----hmdDhtLpgk---ygIGFTnMVerTt-p------gskdls----s
d2c53a- ( 127 ) -amsghGcLekWA-----dGVLLLnTTLTVkrg----------aaasHs-
d2hxma2 ( 184 ) FvhpghGdLsgWAk----qGVLLLnAVLTVrah----------qansHke
d2q07a2 ( 270 ) kpyltSrtHraLrskVkVnvNEIIIssplVVPrefell--------hwse
d4wpka1 ( 107 ) yplPsnGdLtpWAq----rGVLLLnrVLTVrps----------npasHrg
bbbbb
210 220 230 240 250
d1muga- ( 83 ) qeLhaGGrkLiekIedyqPqALAILg-kqAYeqGF----sqr-gaqwgkq
d1oe4a- ( 200 ) tLLeiCdeaLcqAVrvLgVklVIGVg-rfSeqrArkaLmae---------
d1vk2a- ( 100 ) eEqaacghFLlaQIeiInPdVIVALg-atALsfFvd---gkvsitvrGnp
d1wywa- ( 206 ) keFregGriLvqkLqkyqPrIAVFnG-kcIYeiFSkEvfgvkvnLefglQ
d2c53a- ( 163 ) igWdrFVggVIrrLaar-pglVFMLWgthAqnaIp--dp-----------
d2hxma2 ( 220 ) rgWeqFTdaVvswLnqnsngLVFLLWgsyAqkkGsaIdr-----------
d2q07a2 ( 324 ) eeVsfVagwLkrFIekGgFrkVVAHVtggyrkVVerVed-----------
d4wpka1 ( 143 ) kgWeaVTecAIraLaaraapLVAILWgrdAstLkpmLaa-----------
aaaaaaaaaaaaaaa bbbbb aaaaaa
260 270 280 290 300
d1muga- ( 127 ) tltIg--sTqIWVLpnPsglsr--v--slekLveaYreLdqaLvv
d1oe4a- ( 240 ) -----gidVtVkGImhPsprnpqAnkgWegiVrgqLlel-gVlslLt
d1vk2a- ( 148 ) idw-lg-gkkVIPTfhPsyLlrnrsnelrriVleDIekAksfik
d1wywa- ( 256 ) phkIpdteTlCYVMPsSsarcaqfp--rAqDKvHyYikLkdlrdqlkgie
d2c53a- ( 202 ) -------vHcvlkfsnPspls-kvpFgtCqhFlvAnryLet-----sisp
d2hxma2 ( 259 ) ------krHhVlqtahPsplsVyrgFfgcrHFskTnelLqk----sgkkp
d2q07a2 ( 363 ) -----eVeAeVvyTAekdvls--------deSierLkqeIesk-------
d4wpka1 ( 182 ) ------gnCvAIesphPsplsAsrgFfgsrPFsrAnelLvg----mgaep
bbbbb aaaaaaaaaa
310 320
d1muga-
d1oe4a-
d1vk2a-
d1wywa- ( 304 ) rnmdvqevqytfdlqlAqedakkmavk
d2c53a- ( 240 ) IdWsV
d2hxma2 ( 299 ) IdWkel
d2q07a2 ( 393 ) --------------------------g
d4wpka1 ( 222 ) IdWrLp
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| G:T/U mismatch-specific DNA glycosylase, Mug | Mug-like | Escherichia coli [TaxId: 562] | 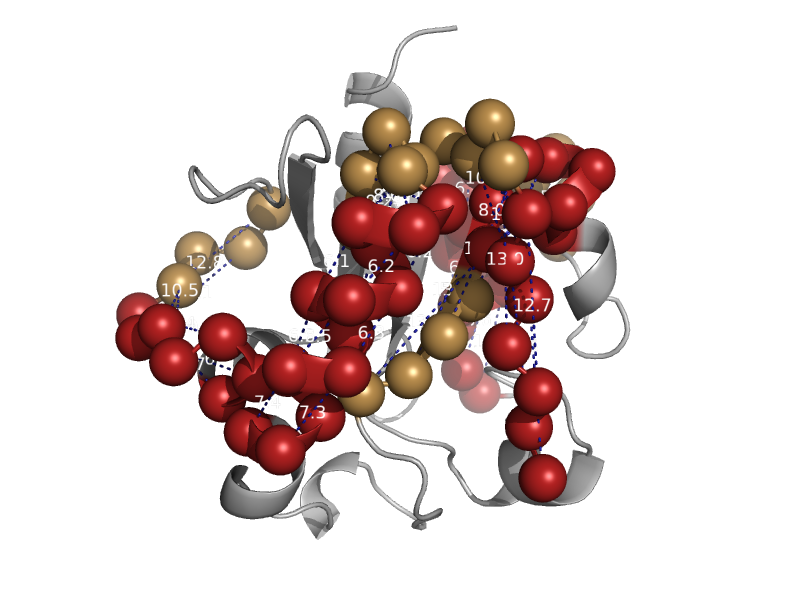 | view | |
| Single-strand selective monofunctional uracil-DNA glycosylase SMUG1 | Single-strand selective monofunctional uracil-DNA glycosylase SMUG1 | African clawed frog (Xenopus laevis) [TaxId: 8355] | 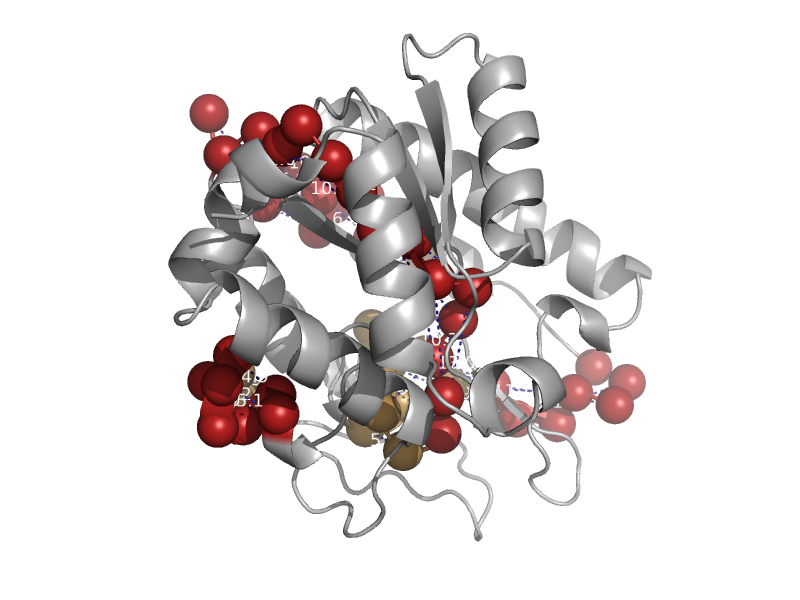 | view | |
| Thermophilic uracil-DNA glycosylase | Mug-like | Thermotoga maritima [TaxId: 2336] | 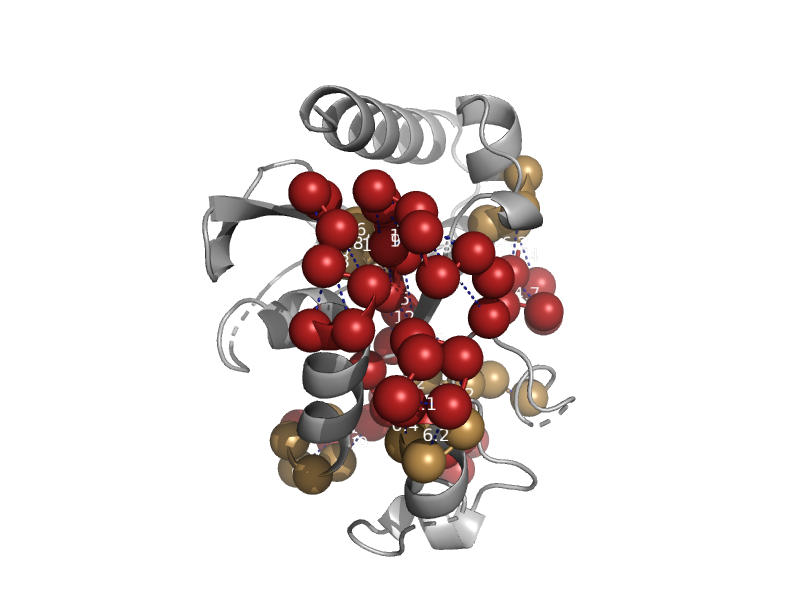 | view | |
| G/T mismatch-specific thymine DNA glycosylase | Mug-like | Human (Homo sapiens) [TaxId: 9606] | 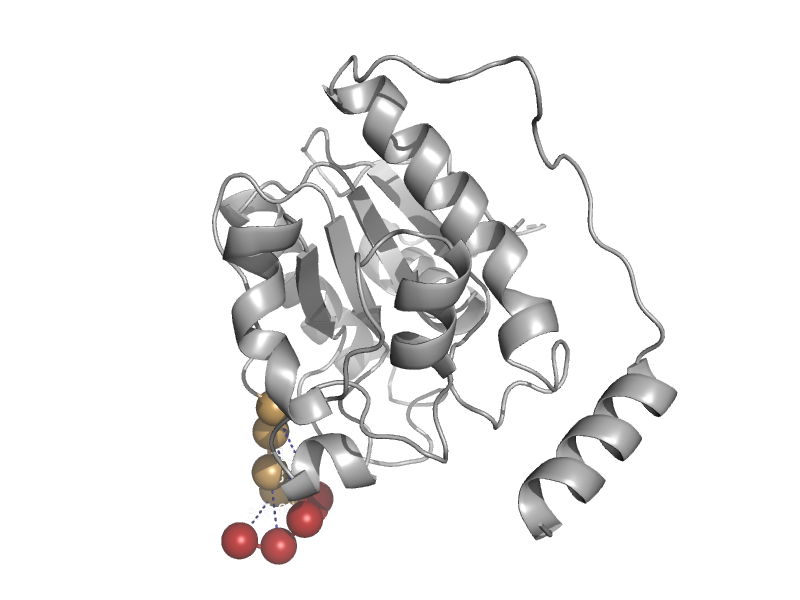 | view | |
| automated matches | Uracil-DNA glycosylase | Human herpesvirus 1 [TaxId: 10298] | 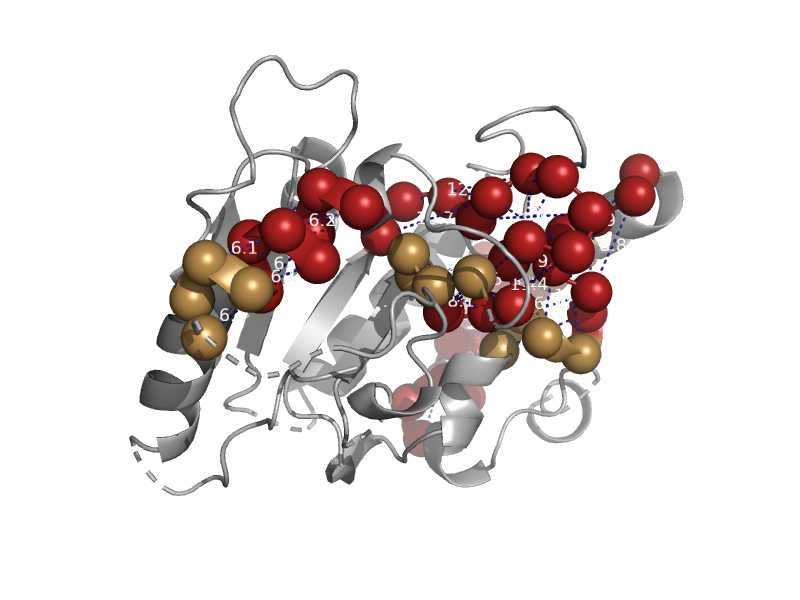 | view | |
| Uracil-DNA glycosylase | Uracil-DNA glycosylase | Human (Homo sapiens) [TaxId: 9606] | 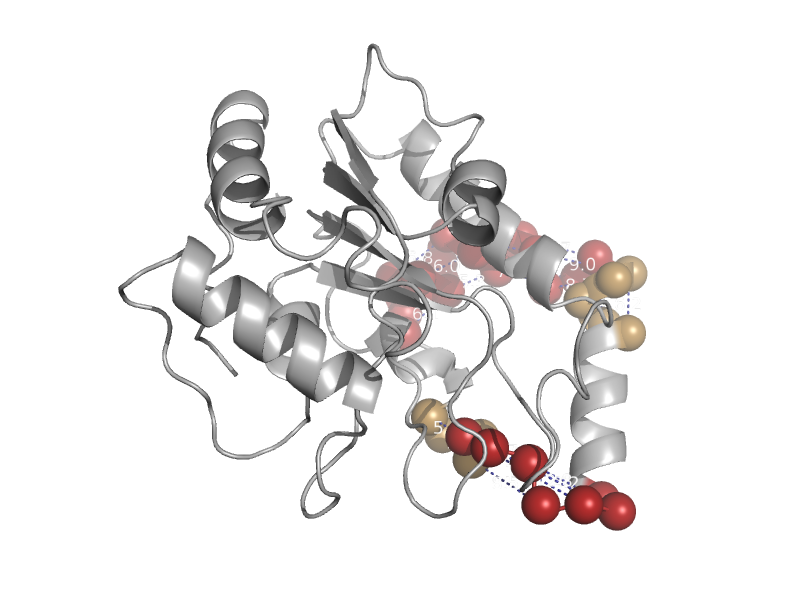 | view | |
| Uncharacterized protein AF0587 | AF0587 domain-like | Archaeoglobus fulgidus [TaxId: 2234] | 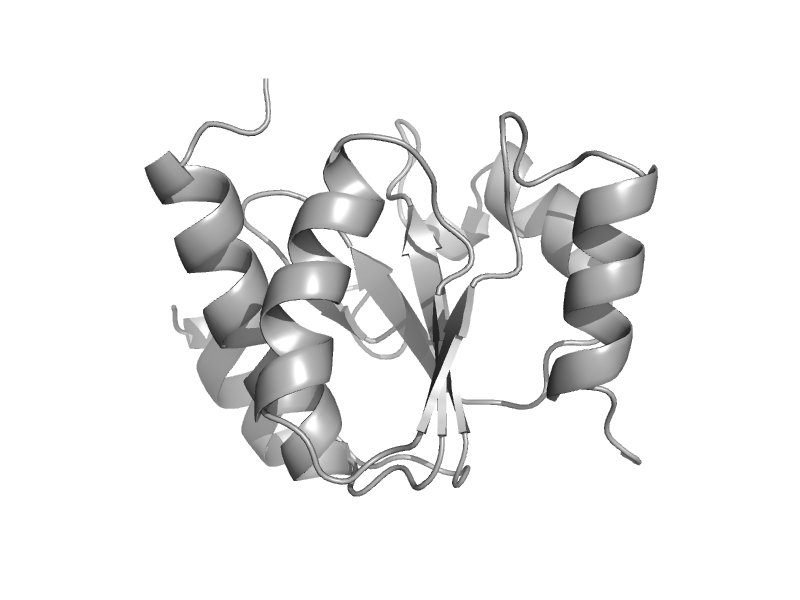 | view | |
| automated matches | automated matches | Mycobacterium tuberculosis [TaxId: 83332] | 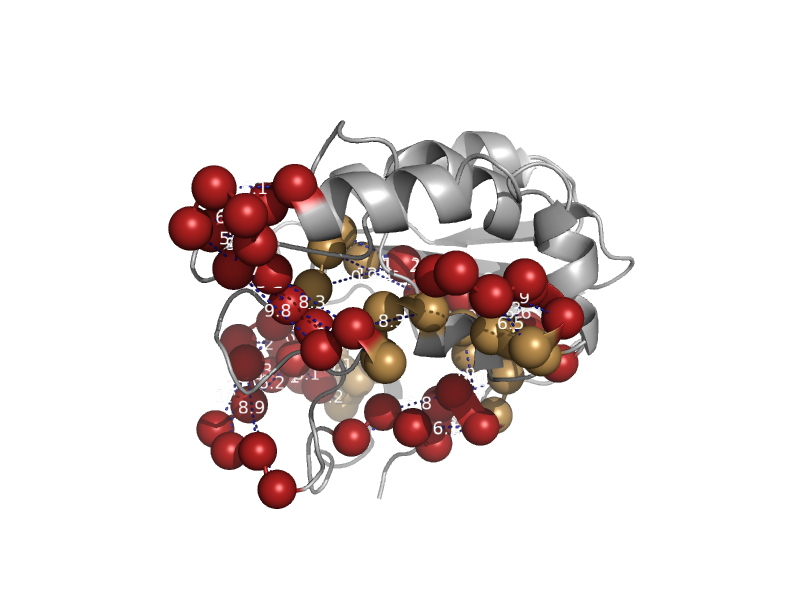 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | V | E | D | I | L | A | P | G | L | R | V | V | F | C | G | I | N | P | G | L | S | - | S | A | G | T | G | F | P | F | A | H | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | N | R | F | W | K | V | I | Y | Q | A | G | - | F | T | D | R | Q | L | K | - | - | - | - | - | - | - | - | - | - | - | P | Q | E | A | Q | H | L | L | - | D | - | - | - | Y | R | C | G | V | T | K | L | V | D | R | P | T | - | V | - | - | - | - | - | - | Q | A | N | E | V | S | - | - | - | - | K | Q | E | L | H | A | G | G | R | K | L | I | E | K | I | E | D | Y | Q | P | Q | A | L | A | I | L | G | - | K | Q | A | Y | E | Q | G | F | - | - | - | - | S | Q | R | - | G | A | Q | W | G | K | Q | T | L | T | I | G | - | - | S | T | Q | I | W | V | L | P | N | P | S | G | L | S | R | - | - | V | - | - | S | L | E | K | L | V | E | A | Y | R | E | L | D | Q | A | L | V | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | E | S | P | A | D | S | F | L | K | V | E | L | E | L | N | L | K | L | S | N | L | V | F | Q | D | P | V | Q | Y | V | Y | N | P | L | V | Y | A | W | A | P | H | E | N | Y | V | Q | T | Y | C | K | S | K | K | E | V | L | F | L | G | M | N | P | G | P | F | G | M | A | Q | T | G | V | P | F | G | E | V | N | H | V | R | D | W | L | Q | I | E | G | P | V | S | K | P | E | V | E | H | P | K | R | R | I | R | G | F | E | C | P | Q | S | E | V | S | G | A | R | F | W | S | L | F | K | S | L | C | - | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | P | E | T | F | F | K | - | - | - | - | - | H | C | F | V | H | N | H | C | P | L | I | F | M | N | H | S | G | K | N | L | T | P | T | D | L | P | K | A | Q | R | D | T | L | L | E | I | C | D | E | A | L | C | Q | A | V | R | V | L | G | V | K | L | V | I | G | V | G | - | R | F | S | E | Q | R | A | R | K | A | L | M | A | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | I | D | V | T | V | K | G | I | M | H | P | S | P | R | N | P | Q | A | N | K | G | W | E | G | I | V | R | G | Q | L | L | E | L | - | G | V | L | S | L | L | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | T | R | E | E | L | E | I | V | S | E | R | V | K | C | T | - | - | - | - | - | - | - | - | - | - | - | - | A | C | P | L | H | L | N | R | T | N | V | V | V | G | E | G | N | L | D | T | R | I | V | F | V | G | E | G | P | G | E | E | - | E | D | - | T | G | R | P | F | V | G | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | G | L | L | T | E | L | L | R | E | S | G | - | - | - | - | - | - | I | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | E | D | - | - | - | - | - | - | - | - | - | V | Y | I | C | N | V | V | K | C | R | P | - | P | - | - | - | - | - | - | N | N | R | T | P | T | - | - | - | - | P | E | E | Q | A | A | C | G | H | F | L | L | A | Q | I | E | I | I | N | P | D | V | I | V | A | L | G | - | A | T | A | L | S | F | F | V | D | - | - | - | G | K | V | S | I | T | V | R | G | N | P | I | D | W | - | L | G | - | G | K | K | V | I | P | T | F | H | P | S | Y | L | L | R | N | R | S | N | E | L | R | R | I | V | L | E | D | I | E | K | A | K | S | F | I | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | E | L | L | T | K | T | L | P | D | I | L | T | F | N | L | D | I | V | I | I | G | I | N | P | G | L | M | - | A | A | Y | K | G | H | H | Y | P | G | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | N | H | F | W | K | C | L | F | M | S | G | - | L | S | E | V | Q | L | N | - | - | - | - | - | - | - | - | - | - | - | H | M | D | D | H | T | L | P | G | K | - | - | - | Y | G | I | G | F | T | N | M | V | E | R | T | T | - | P | - | - | - | - | - | - | G | S | K | D | L | S | - | - | - | - | S | K | E | F | R | E | G | G | R | I | L | V | Q | K | L | Q | K | Y | Q | P | R | I | A | V | F | N | G | - | K | C | I | Y | E | I | F | S | K | E | V | F | G | V | K | V | N | L | E | F | G | L | Q | P | H | K | I | P | D | T | E | T | L | C | Y | V | M | P | S | S | S | A | R | C | A | Q | F | P | - | - | R | A | Q | D | K | V | H | Y | Y | I | K | L | K | D | L | R | D | Q | L | K | G | I | E | R | N | M | D | V | Q | E | V | Q | Y | T | F | D | L | Q | L | A | Q | E | D | A | K | K | M | A | V | K |
| D | W | T | T | F | R | V | F | L | I | D | D | A | W | R | P | L | M | E | P | E | L | A | N | P | L | T | A | H | L | L | A | E | Y | N | R | R | C | Q | - | - | T | E | E | V | L | P | P | R | E | D | V | F | S | W | T | R | Y | C | T | P | D | E | V | R | V | V | I | I | G | Q | N | P | Y | H | H | P | G | Q | A | H | G | L | A | F | S | V | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | V | - | P | P | P | P | S | L | R | N | V | L | A | A | V | K | N | C | Y | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | M | S | G | H | G | C | L | E | K | W | A | - | - | - | - | - | D | G | V | L | L | L | N | T | T | L | T | V | K | R | G | - | - | - | - | - | - | - | - | - | - | A | A | A | S | H | S | - | I | G | W | D | R | F | V | G | G | V | I | R | R | L | A | A | R | - | P | G | L | V | F | M | L | W | G | T | H | A | Q | N | A | I | P | - | - | D | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V | H | C | V | L | K | F | S | N | P | S | P | L | S | - | K | V | P | F | G | T | C | Q | H | F | L | V | A | N | R | Y | L | E | T | - | - | - | - | - | S | I | S | P | I | D | W | S | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | F | G | E | S | W | K | K | H | L | S | G | E | F | G | K | P | Y | F | I | K | L | M | G | F | V | A | E | E | R | K | - | - | H | Y | T | V | Y | P | P | P | H | Q | V | F | T | W | T | Q | M | C | D | I | K | D | V | K | V | V | I | L | G | Q | D | P | Y | H | G | P | N | Q | A | H | G | L | C | F | S | V | Q | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | V | - | P | P | P | P | S | L | E | N | I | Y | K | E | L | S | T | D | I | E | - | - | - | - | - | - | - | - | - | - | - | - | D | F | V | H | P | G | H | G | D | L | S | G | W | A | K | - | - | - | - | Q | G | V | L | L | L | N | A | V | L | T | V | R | A | H | - | - | - | - | - | - | - | - | - | - | Q | A | N | S | H | K | E | R | G | W | E | Q | F | T | D | A | V | V | S | W | L | N | Q | N | S | N | G | L | V | F | L | L | W | G | S | Y | A | Q | K | K | G | S | A | I | D | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | R | H | H | V | L | Q | T | A | H | P | S | P | L | S | V | Y | R | G | F | F | G | C | R | H | F | S | K | T | N | E | L | L | Q | K | - | - | - | - | S | G | K | K | P | I | D | W | K | E | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | Y | F | F | E | R | A | L | E | C | Y | K | P | F | S | D | T | V | L | L | L | P | C | T | A | R | K | P | Y | L | T | S | R | T | H | R | A | L | R | S | K | V | K | V | N | V | N | E | I | I | I | S | S | P | L | V | V | P | R | E | F | E | L | L | - | - | - | - | - | - | - | - | H | W | S | E | E | E | V | S | F | V | A | G | W | L | K | R | F | I | E | K | G | G | F | R | K | V | V | A | H | V | T | G | G | Y | R | K | V | V | E | R | V | E | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | V | E | A | E | V | V | Y | T | A | E | K | D | V | L | S | - | - | - | - | - | - | - | - | D | E | S | I | E | R | L | K | Q | E | I | E | S | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G |
| - | M | A | R | P | L | S | E | L | V | E | R | G | W | A | A | A | L | E | P | V | A | - | - | D | Q | V | A | H | M | G | Q | F | L | R | A | E | I | A | - | A | G | R | R | Y | L | P | A | G | S | N | V | L | R | A | F | T | - | F | P | F | D | N | V | R | V | L | I | V | G | Q | D | P | Y | P | T | P | G | H | A | V | G | L | S | F | S | V | A | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | V | R | P | W | P | R | S | L | A | N | I | F | D | E | Y | T | A | D | L | G | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | P | L | P | S | N | G | D | L | T | P | W | A | Q | - | - | - | - | R | G | V | L | L | L | N | R | V | L | T | V | R | P | S | - | - | - | - | - | - | - | - | - | - | N | P | A | S | H | R | G | K | G | W | E | A | V | T | E | C | A | I | R | A | L | A | A | R | A | A | P | L | V | A | I | L | W | G | R | D | A | S | T | L | K | P | M | L | A | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | N | C | V | A | I | E | S | P | H | P | S | P | L | S | A | S | R | G | F | F | G | S | R | P | F | S | R | A | N | E | L | L | V | G | - | - | - | - | M | G | A | E | P | I | D | W | R | L | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1muga_ | A | P0A9H1 | GO:0000700 |
| d1muga_ | A | P0A9H1 | GO:0003677 |
| d1muga_ | A | P0A9H1 | GO:0004844 |
| d1muga_ | A | P0A9H1 | GO:0005737 |
| d1muga_ | A | P0A9H1 | GO:0006281 |
| d1muga_ | A | P0A9H1 | GO:0006285 |
| d1muga_ | A | P0A9H1 | GO:0008263 |
| d1muga_ | A | P0A9H1 | GO:0016787 |
| d1muga_ | A | P0A9H1 | GO:0043739 |
| d1oe4a_ | A | Q9YGN6 | GO:0006284 |
| d1oe4a_ | A | Q9YGN6 | GO:0017065 |
| d1vk2a_ | A | Q9WYY1 | GO:0004844 |
| d1vk2a_ | A | Q9WYY1 | GO:0006281 |
| d1vk2a_ | A | Q9WYY1 | GO:0016787 |
| d1vk2a_ | A | Q9WYY1 | GO:0046872 |
| d1vk2a_ | A | Q9WYY1 | GO:0051539 |
| d2q07a2 | A | O29668 | GO:0003723 |
| d2q07a2 | A | O29668 | GO:0003723 |
