solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1glaf- ( 1 ) gfdlslv-tieIiAPL
d1gpra- ( 4 ) eplqneigeevfVSPI
d1qwya- ( 45 ) vsygtyytidsngdyHHTpdgnwnqamfdnkeysYtFvdaqghtHyFYn-
60 70 80 90 100
d1glaf- ( 27 ) sgeIvnIedVpd---vvfaekivgdGIAIkPt----------gnkMVAPV
d1gpra- ( 20 ) tgeIhpItdVpd---qvfsgkmmgdGFAIlPs----------eGiVvSPV
d1qwya- ( 94 ) -cyPknAnangsgqtyVnpatagDnNDYTaSQsqqhinqygYqsnvGpDa
110 120 130 140 150
d1glaf- ( 64 ) d---------------gtIgkIfe------tnHAFsIeSdsgVeLfVhFG
d1gpra- ( 57 ) r---------------GkIlnvfp------tkHAIgLqSdggreIlIhFG
d1qwya- ( 143 ) syyssghAkdaswLtsrkqlqpygqYhgggaHyGVDYaMpenspVySLTd
bbb bbbb bbb
160 170 180 190 200
d1glaf- ( 93 ) idTveLkgeGFkriAeegqrV-vgdtVIeFdlplLeekA-stlTPVVIsn
d1gpra- ( 86 ) IdTvslkgegFtsfVsegdrVepGqkLLeVdldaVkpnVpslmTPIVFtn
d1qwya- ( 229 ) ----------gtVvqAgWsnyGGGNqVTIkea-------nsnnYQWYMHN
bbb bbbbb bbb
210 220 230 240 250
d1glaf- ( 143 ) mdeIkeLik-lsgsVtvgetpVIrIkk
d1gpra- ( 136 ) laegetVsikasgsVnreqedIVkie
d1qwya- ( 262 ) n----rLtvsagdkVkag-dqIAySGstGnStapHVHFQrMsgGIGNqyA
bb bb bb
d1glaf-
d1gpra-
d1qwya- ( 307 ) vdPtsYLq
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Glucose-specific factor III (glsIII) | Glucose permease-like | Escherichia coli [TaxId: 562] | 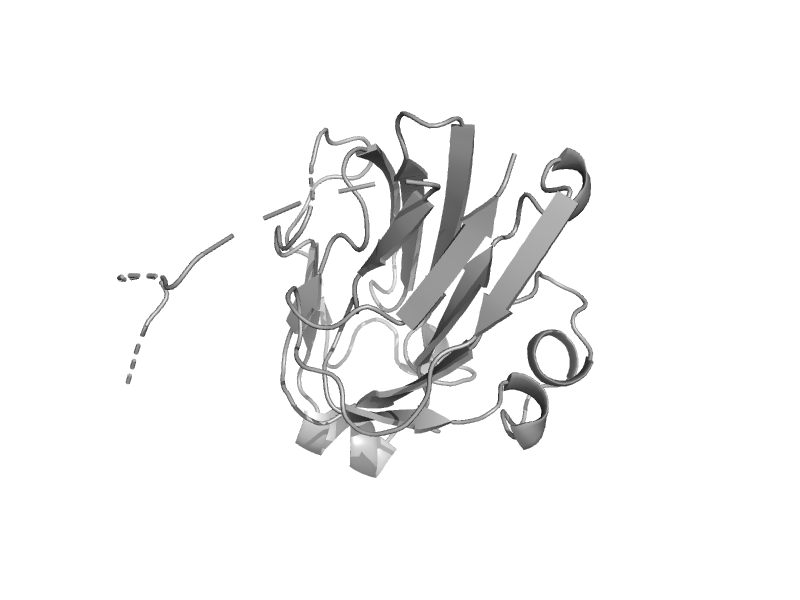 | view | |
| Glucose permease IIa domain, IIa-glc | Glucose permease-like | Bacillus subtilis [TaxId: 1423] | 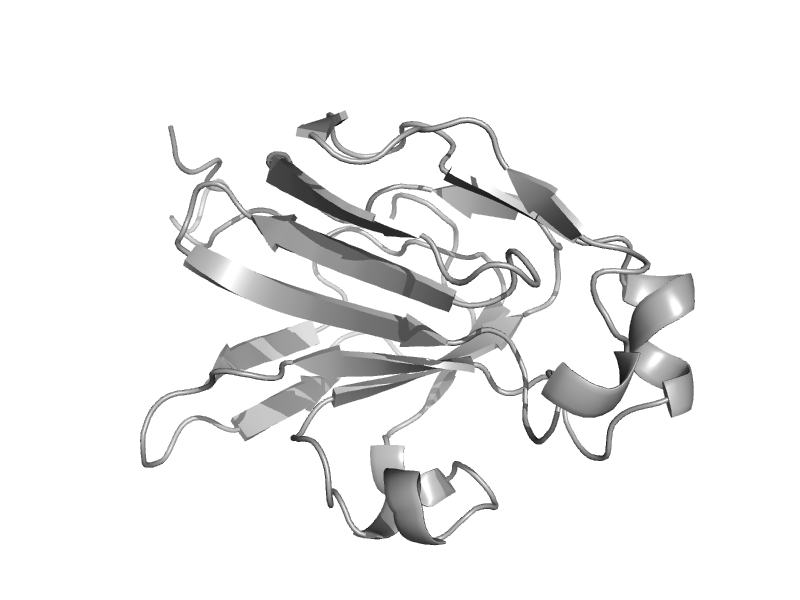 | view | |
| Peptidoglycan hydrolase LytM | Peptidoglycan hydrolase LytM | Staphylococcus aureus [TaxId: 1280] | 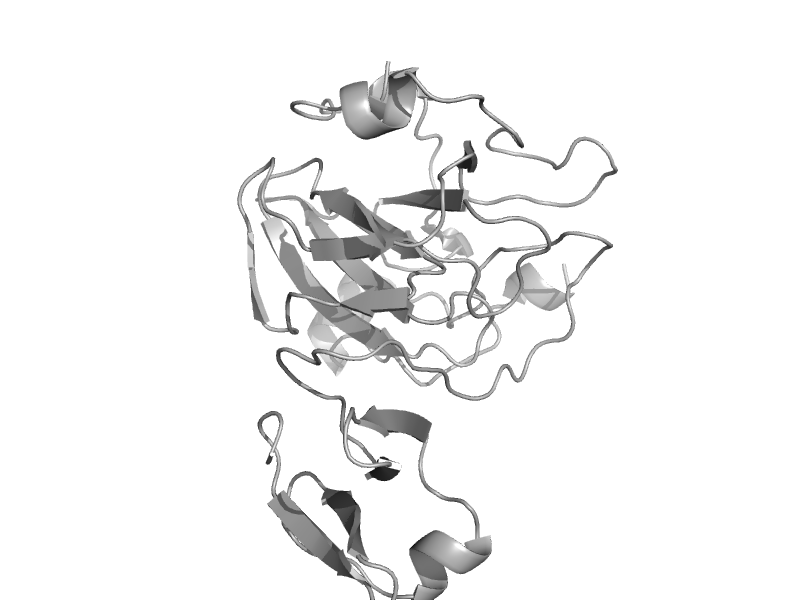 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | F | D | L | S | L | V | - | T | I | E | I | I | A | P | L | S | G | E | I | V | N | I | E | D | V | P | D | - | - | - | V | V | F | A | E | K | I | V | G | D | G | I | A | I | K | P | T | - | - | - | - | - | - | - | - | - | - | G | N | K | M | V | A | P | V | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | T | I | G | K | I | F | E | - | - | - | - | - | - | T | N | H | A | F | S | I | E | S | D | S | G | V | E | L | F | V | H | F | G | I | D | T | V | E | L | K | G | E | G | F | K | R | I | A | E | E | G | Q | R | V | - | V | G | D | T | V | I | E | F | D | L | P | L | L | E | E | K | A | - | S | T | L | T | P | V | V | I | S | N | M | D | E | I | K | E | L | I | K | - | L | S | G | S | V | T | V | G | E | T | P | V | I | R | I | K | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | P | L | Q | N | E | I | G | E | E | V | F | V | S | P | I | T | G | E | I | H | P | I | T | D | V | P | D | - | - | - | Q | V | F | S | G | K | M | M | G | D | G | F | A | I | L | P | S | - | - | - | - | - | - | - | - | - | - | E | G | I | V | V | S | P | V | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | K | I | L | N | V | F | P | - | - | - | - | - | - | T | K | H | A | I | G | L | Q | S | D | G | G | R | E | I | L | I | H | F | G | I | D | T | V | S | L | K | G | E | G | F | T | S | F | V | S | E | G | D | R | V | E | P | G | Q | K | L | L | E | V | D | L | D | A | V | K | P | N | V | P | S | L | M | T | P | I | V | F | T | N | L | A | E | G | E | T | V | S | I | K | A | S | G | S | V | N | R | E | Q | E | D | I | V | K | I | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| V | S | Y | G | T | Y | Y | T | I | D | S | N | G | D | Y | H | H | T | P | D | G | N | W | N | Q | A | M | F | D | N | K | E | Y | S | Y | T | F | V | D | A | Q | G | H | T | H | Y | F | Y | N | - | - | C | Y | P | K | N | A | N | A | N | G | S | G | Q | T | Y | V | N | P | A | T | A | G | D | N | N | D | Y | T | A | S | Q | S | Q | Q | H | I | N | Q | Y | G | Y | Q | S | N | V | G | P | D | A | S | Y | Y | S | S | G | H | A | K | D | A | S | W | L | T | S | R | K | Q | L | Q | P | Y | G | Q | Y | H | G | G | G | A | H | Y | G | V | D | Y | A | M | P | E | N | S | P | V | Y | S | L | T | D | - | - | - | - | - | - | - | - | - | - | G | T | V | V | Q | A | G | W | S | N | Y | G | G | G | N | Q | V | T | I | K | E | A | - | - | - | - | - | - | - | N | S | N | N | Y | Q | W | Y | M | H | N | N | - | - | - | - | R | L | T | V | S | A | G | D | K | V | K | A | G | - | D | Q | I | A | Y | S | G | S | T | G | N | S | T | A | P | H | V | H | F | Q | R | M | S | G | G | I | G | N | Q | Y | A | V | D | P | T | S | Y | L | Q |
