solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1kb0a2 ( 1 ) tgpaaqAaaAVqrVdgdfIranaarTpdWPtiGVdyaEtRYSrldqInaa
d2ad6a- ( 1 ) dadLdkqvntaGAWpiaTGgyySqHnsplaqInks
d4cvba- ( 45 ) saienfqpVtaddLa--gknpaNWPIlRGnYqGwGySpldqInkd
d6zcwa- ( 28 ) aVsneeIlqDpknpqqIVtnGLgvqGqRySpldlLnvn
aaaaaa bb
60 70 80 90 100
d1kb0a2 ( 51 ) nVkdlgLawsynLe--strGVEATPVVVdGIMYVSASw-SvVHAIDTrtG
d2ad6a- ( 36 ) nVknVkaawsfsTg--vlnGHEgAPLVigdmMYVHSAfpNNTYALnLndp
d4cvba- ( 88 ) nVgdLqlvwsrtMe---pgsNEGAAIAYngVIFLGNTn-DVIQAIdGktG
d6zcwa- ( 66 ) nVkeLrpvwafsFggqkqrGQQAQPLIkdgvMYLTGSy-SRVFAVdArtG
333 bbbbbbb bbb bbbbb bbbbb
110 120 130 140 150
d1kb0a2 ( 98 ) -nrIwtydPqIdrstgfkgccDVVNrGVaLWkgkVYVGAWdGrLIALdAa
d2ad6a- ( 84 ) gkivwqhkPkQdasTkavMccDVVDRGLaYGagqIVKKQAnGhLLALdak
d4cvba- ( 134 ) -sliweyrrklpsaskFiNslGaaKRSIALfgdkVYFVSwdNfVVALdAk
d6zcwa- ( 115 ) -kklwqydArlpddi--rpccdvINrGVaLygnlVFFGTLDAkLVALnkd
bbbbb 333 bbb bbbbb bbbbbb
160 170 180 190 200
d1kb0a2 ( 147 ) tGkevwhqnTfegqkgsLTItgAPrVFkgkVIIGNggAeYGVrGYITAYd
d2ad6a- ( 134 ) tGkinweveVÇd-pkvGsTLtQAPfVAkdTVLMGçsgAeLGvrGaVNAFd
d4cvba- ( 183 ) tGklawetnRgqGveeGVANSSGPIVVdgVVIAGSTÇqfSgfGÇYVTGTd
d6zcwa- ( 162 ) tGkvvwskkVad-hkeGySIsAAPmIVngkLITGVagGefGVvGkIQAYn
bbbbbb 333 bbb bbbb bbbbbb
210 220 230 240 250
d1kb0a2 ( 197 ) AetGerkwRWfSVPgdps--------kpfed------eSMkrAartWdps
d2ad6a- ( 183 ) LktGelkwrAfATGsddsVrLa-----kdFNsanphYgqfglGtktWeg-
d4cvba- ( 233 ) AesGeelwrntFIprpg-----------------------eegddtWggA
d6zcwa- ( 211 ) PenGellWmRPTVEGh--mGyvykdgkaiengiSg-----geagktWpg-
bbbbbb
260 270 280 290 300
d1kb0a2 ( 233 ) gkWweAgGGGTMwdSMTFdaelnTMYVGTGNGSPWShkvRSpkggdNLYL
d2ad6a- ( 227 ) -dawk-iGGGTNwGWYAYDpklnLFYYGSGNPAPWNetmRp---gdNkWT
d4cvba- ( 260 ) p--yenRwMTGAwGQITYDpeldLVYYGSTGAGPASEvqRgTeggtLagT
d6zcwa- ( 253 ) -dlwk-tGGAAPwlggyYDpetnlILFGTGNPAPWNShlRp---gdNlyS
33 bbb bbbbb
310 320 330 340 350
d1kb0a2 ( 283 ) ASIVALdPdtGkykwhyQETpgDnWDYTSTQpMILadikia---------
d2ad6a- ( 272 ) MTIWGRdLdtGmAkWGYQKTphDeWDFAGVNQMVLTdQpvn---------
d4cvba- ( 308 ) NTRfAVkPktGevvwkhqTLprDNWdSEÇTFEMMVVsTsVnPdakAdgMm
d6zcwa- ( 298 ) SSRLALnPddGtikwhfqSTPhDGWDFDGVNeLISFnykdg---------
bbbbb bbbbbb bbbbbb
360 370 380 390 400
d1kb0a2 ( 324 ) --------gkprkVILHAP-KNGFFFVLdrtnGkfisAknFV-pVnWAsg
d2ad6a- ( 313 ) --------gkmtpLLSHID-RNGILYTLnRenGnLivAekVDpaVnVFkk
d4cvba- ( 358 ) svGanvprgetrkVLTGVPÇKTGVAWQFdAktGdYFWSKaTV-eqnSIas
d6zcwa- ( 339 ) --------gkevkAAATAD-RNGFFYVLdRtnGkfirGfpFVdkItWAtg
bbbbbbb bbbbbb bbbbbb bbb
410 420 430 440 450
d1kb0a2 ( 364 ) Yd-khGkpigiaaardgs-------kpqdAvPGpyGAHNWHPMSFNpqtg
d2ad6a- ( 354 ) VdlktGtPvrdpefaTrmd-----hkGtnIÇPSAMGFHNQGVDSYDpesr
d4cvba- ( 407 ) Id-dtGlVtVnedmILke-----pgktYnYçPTflGGRDWPSAGYLpksn
d6zcwa- ( 380 ) Ld-kdGrPiyndasrPgapgseakgssvfVAPAFLGAKnWMPMAyNkdtg
b bbb 333 bbb bbb
460 470 480 490 500
d1kb0a2 ( 406 ) lVYLPAQNvpVNLmDDkkwefnqagpgkpqsgtgwNTAKffnaeppk---
d2ad6a- ( 399 ) TLYAGLNHIÇMdWePf-mlpyr---------agqfFvGAtLaMypGpnGp
d4cvba- ( 451 ) LYVIPLSNAÇydVmArtt-----------eAtpadvyNTdAtLvlAp---
d6zcwa- ( 429 ) lFYVPSNEWGMdIwNe-giayk---------kGaaFlGAgftIkplne--
bbbbbbb bbbbbbb bbbbbbb
510 520 530 540 550
d1kb0a2 ( 453 ) -skpfGrLLAWdPvaqkaawsvehvspWNGGTLTTaGnVVFQGTAdGrLV
d2ad6a- ( 439 ) tkkeMGQIrAFdLTtGkakwtkwekFAAWGGTLYTkggLVWYATLdGyLk
d4cvba- ( 487 ) gktnMGrVDAIdLatGetkwsyetrAALYDPVLTTGgdLVFVGGIdrdFR
d6zcwa- ( 467 ) --dyIGVLRAIdPvsgkevwrhknyAPLWGGVLTTkgnLVFTGTPeGfLQ
bbbbbbb bbbbbbb bbbb bbbbb bbb
560 570 580 590 600
d1kb0a2 ( 502 ) AYhAatGekl-eapTgtGVVAAPSTYmvdgrQyVSVAVgwGGvyGlaara
d2ad6a- ( 489 ) ALdnkdGkelwnfkMpSGGIgSPMTYsfkgkQyIGSMyGVGGwPGvglVf
d4cvba- ( 537 ) ALdAesGkevwstrLpGAVSGYTTSYsidgrQYVAVVSG-GSl-------
d6zcwa- ( 515 ) AFnAktGdkvwefqTgsGVLGSPVTWeMdgeQYVSVVSGWgGaVp-----
bbb bbbbbb bbbbb bbbbbbbb
610 620 630 640
d1kb0a2 ( 552 ) te-------------------rqgpGtVYTFVvggkarmpe
d2ad6a- ( 539 ) dltdpsaglGAVgAFreLqnhtqmgGGLmVFSl
d4cvba- ( 579 ) ------GGptFGptTpdv-dsasganGIyVFAlpe
d6zcwa- ( 560 ) --------lwGgeVakrVkd-fnqgGmLwTFkLp--kqlqq
bbbbbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Quinoprotein alcohol dehydrogenase, N-terminal domain | Quinoprotein alcohol dehydrogenase-like | Comamonas testosteroni [TaxId: 285] | 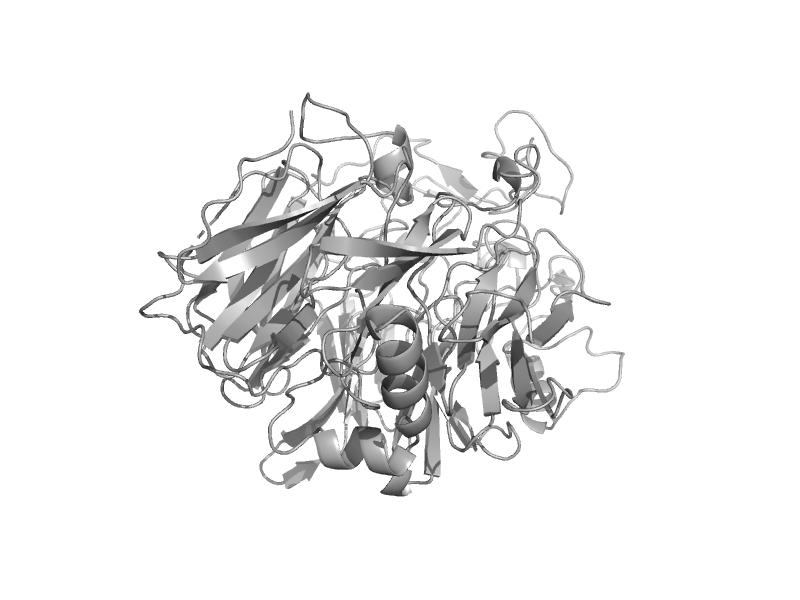 | view | |
| Methanol dehydrogenase, heavy chain | Quinoprotein alcohol dehydrogenase-like | Methylophilus methylotrophus, w3a1 [TaxId: 17] | 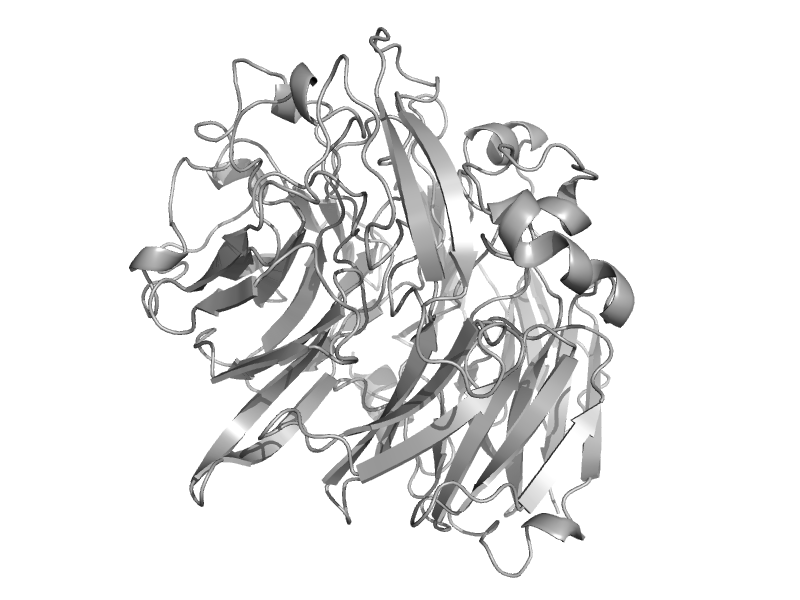 | view | |
| automated matches | automated matches | Pseudogluconobacter saccharoketogenes [TaxId: 133921] | 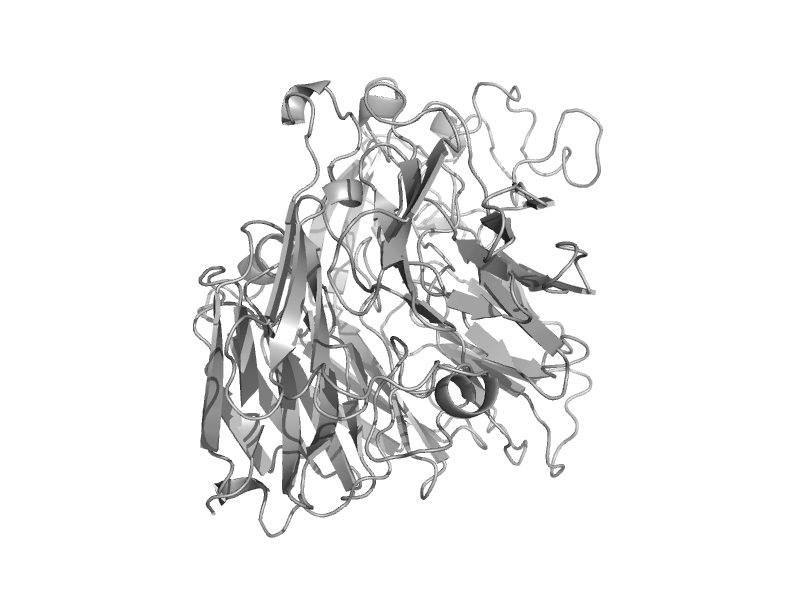 | view | |
| automated matches | automated matches | Pseudomonas putida [TaxId: 160488] | 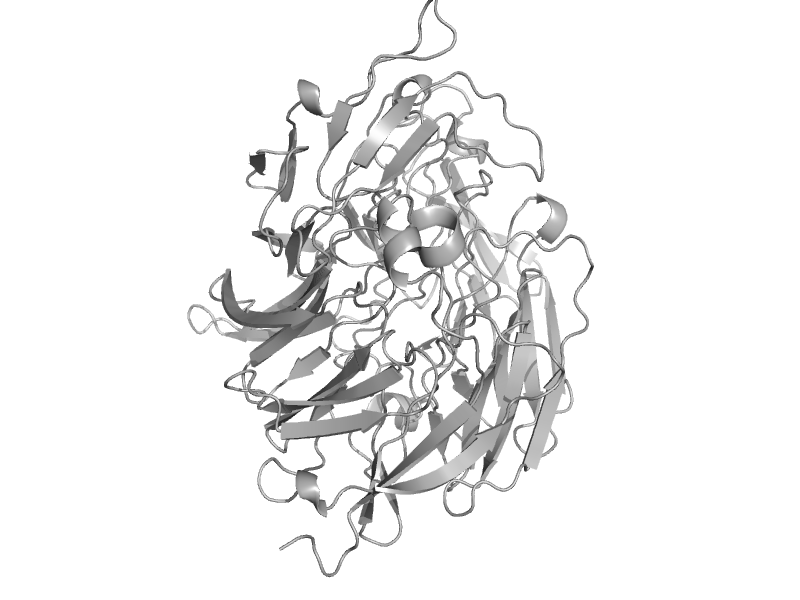 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | 330 | 340 | 350 | 360 | 370 | 380 | 390 | 400 | 410 | 420 | 430 | 440 | 450 | 460 | 470 | 480 | 490 | 500 | 510 | 520 | 530 | 540 | 550 | 560 | 570 | 580 | 590 | 600 | 610 | 620 | 630 | 640 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | G | P | A | A | Q | A | A | A | A | V | Q | R | V | D | G | D | F | I | R | A | N | A | A | R | T | P | D | W | P | T | I | G | V | D | Y | A | E | T | R | Y | S | R | L | D | Q | I | N | A | A | N | V | K | D | L | G | L | A | W | S | Y | N | L | E | - | - | S | T | R | G | V | E | A | T | P | V | V | V | D | G | I | M | Y | V | S | A | S | W | - | S | V | V | H | A | I | D | T | R | T | G | - | N | R | I | W | T | Y | D | P | Q | I | D | R | S | T | G | F | K | G | C | C | D | V | V | N | R | G | V | A | L | W | K | G | K | V | Y | V | G | A | W | D | G | R | L | I | A | L | D | A | A | T | G | K | E | V | W | H | Q | N | T | F | E | G | Q | K | G | S | L | T | I | T | G | A | P | R | V | F | K | G | K | V | I | I | G | N | G | G | A | E | Y | G | V | R | G | Y | I | T | A | Y | D | A | E | T | G | E | R | K | W | R | W | F | S | V | P | G | D | P | S | - | - | - | - | - | - | - | - | K | P | F | E | D | - | - | - | - | - | - | E | S | M | K | R | A | A | R | T | W | D | P | S | G | K | W | W | E | A | G | G | G | G | T | M | W | D | S | M | T | F | D | A | E | L | N | T | M | Y | V | G | T | G | N | G | S | P | W | S | H | K | V | R | S | P | K | G | G | D | N | L | Y | L | A | S | I | V | A | L | D | P | D | T | G | K | Y | K | W | H | Y | Q | E | T | P | G | D | N | W | D | Y | T | S | T | Q | P | M | I | L | A | D | I | K | I | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | K | P | R | K | V | I | L | H | A | P | - | K | N | G | F | F | F | V | L | D | R | T | N | G | K | F | I | S | A | K | N | F | V | - | P | V | N | W | A | S | G | Y | D | - | K | H | G | K | P | I | G | I | A | A | A | R | D | G | S | - | - | - | - | - | - | - | K | P | Q | D | A | V | P | G | P | Y | G | A | H | N | W | H | P | M | S | F | N | P | Q | T | G | L | V | Y | L | P | A | Q | N | V | P | V | N | L | M | D | D | K | K | W | E | F | N | Q | A | G | P | G | K | P | Q | S | G | T | G | W | N | T | A | K | F | F | N | A | E | P | P | K | - | - | - | - | S | K | P | F | G | R | L | L | A | W | D | P | V | A | Q | K | A | A | W | S | V | E | H | V | S | P | W | N | G | G | T | L | T | T | A | G | N | V | V | F | Q | G | T | A | D | G | R | L | V | A | Y | H | A | A | T | G | E | K | L | - | E | A | P | T | G | T | G | V | V | A | A | P | S | T | Y | M | V | D | G | R | Q | Y | V | S | V | A | V | G | W | G | G | V | Y | G | L | A | A | R | A | T | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | Q | G | P | G | T | V | Y | T | F | V | V | G | G | K | A | R | M | P | E |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | A | D | L | D | K | Q | V | N | T | A | G | A | W | P | I | A | T | G | G | Y | Y | S | Q | H | N | S | P | L | A | Q | I | N | K | S | N | V | K | N | V | K | A | A | W | S | F | S | T | G | - | - | V | L | N | G | H | E | G | A | P | L | V | I | G | D | M | M | Y | V | H | S | A | F | P | N | N | T | Y | A | L | N | L | N | D | P | G | K | I | V | W | Q | H | K | P | K | Q | D | A | S | T | K | A | V | M | C | C | D | V | V | D | R | G | L | A | Y | G | A | G | Q | I | V | K | K | Q | A | N | G | H | L | L | A | L | D | A | K | T | G | K | I | N | W | E | V | E | V | C | D | - | P | K | V | G | S | T | L | T | Q | A | P | F | V | A | K | D | T | V | L | M | G | C | S | G | A | E | L | G | V | R | G | A | V | N | A | F | D | L | K | T | G | E | L | K | W | R | A | F | A | T | G | S | D | D | S | V | R | L | A | - | - | - | - | - | K | D | F | N | S | A | N | P | H | Y | G | Q | F | G | L | G | T | K | T | W | E | G | - | - | D | A | W | K | - | I | G | G | G | T | N | W | G | W | Y | A | Y | D | P | K | L | N | L | F | Y | Y | G | S | G | N | P | A | P | W | N | E | T | M | R | P | - | - | - | G | D | N | K | W | T | M | T | I | W | G | R | D | L | D | T | G | M | A | K | W | G | Y | Q | K | T | P | H | D | E | W | D | F | A | G | V | N | Q | M | V | L | T | D | Q | P | V | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | K | M | T | P | L | L | S | H | I | D | - | R | N | G | I | L | Y | T | L | N | R | E | N | G | N | L | I | V | A | E | K | V | D | P | A | V | N | V | F | K | K | V | D | L | K | T | G | T | P | V | R | D | P | E | F | A | T | R | M | D | - | - | - | - | - | H | K | G | T | N | I | C | P | S | A | M | G | F | H | N | Q | G | V | D | S | Y | D | P | E | S | R | T | L | Y | A | G | L | N | H | I | C | M | D | W | E | P | F | - | M | L | P | Y | R | - | - | - | - | - | - | - | - | - | A | G | Q | F | F | V | G | A | T | L | A | M | Y | P | G | P | N | G | P | T | K | K | E | M | G | Q | I | R | A | F | D | L | T | T | G | K | A | K | W | T | K | W | E | K | F | A | A | W | G | G | T | L | Y | T | K | G | G | L | V | W | Y | A | T | L | D | G | Y | L | K | A | L | D | N | K | D | G | K | E | L | W | N | F | K | M | P | S | G | G | I | G | S | P | M | T | Y | S | F | K | G | K | Q | Y | I | G | S | M | Y | G | V | G | G | W | P | G | V | G | L | V | F | D | L | T | D | P | S | A | G | L | G | A | V | G | A | F | R | E | L | Q | N | H | T | Q | M | G | G | G | L | M | V | F | S | L | - | - | - | - | - | - | - | - |
| - | - | - | - | - | S | A | I | E | N | F | Q | P | V | T | A | D | D | L | A | - | - | G | K | N | P | A | N | W | P | I | L | R | G | N | Y | Q | G | W | G | Y | S | P | L | D | Q | I | N | K | D | N | V | G | D | L | Q | L | V | W | S | R | T | M | E | - | - | - | P | G | S | N | E | G | A | A | I | A | Y | N | G | V | I | F | L | G | N | T | N | - | D | V | I | Q | A | I | D | G | K | T | G | - | S | L | I | W | E | Y | R | R | K | L | P | S | A | S | K | F | I | N | S | L | G | A | A | K | R | S | I | A | L | F | G | D | K | V | Y | F | V | S | W | D | N | F | V | V | A | L | D | A | K | T | G | K | L | A | W | E | T | N | R | G | Q | G | V | E | E | G | V | A | N | S | S | G | P | I | V | V | D | G | V | V | I | A | G | S | T | C | Q | F | S | G | F | G | C | Y | V | T | G | T | D | A | E | S | G | E | E | L | W | R | N | T | F | I | P | R | P | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | E | G | D | D | T | W | G | G | A | P | - | - | Y | E | N | R | W | M | T | G | A | W | G | Q | I | T | Y | D | P | E | L | D | L | V | Y | Y | G | S | T | G | A | G | P | A | S | E | V | Q | R | G | T | E | G | G | T | L | A | G | T | N | T | R | F | A | V | K | P | K | T | G | E | V | V | W | K | H | Q | T | L | P | R | D | N | W | D | S | E | C | T | F | E | M | M | V | V | S | T | S | V | N | P | D | A | K | A | D | G | M | M | S | V | G | A | N | V | P | R | G | E | T | R | K | V | L | T | G | V | P | C | K | T | G | V | A | W | Q | F | D | A | K | T | G | D | Y | F | W | S | K | A | T | V | - | E | Q | N | S | I | A | S | I | D | - | D | T | G | L | V | T | V | N | E | D | M | I | L | K | E | - | - | - | - | - | P | G | K | T | Y | N | Y | C | P | T | F | L | G | G | R | D | W | P | S | A | G | Y | L | P | K | S | N | L | Y | V | I | P | L | S | N | A | C | Y | D | V | M | A | R | T | T | - | - | - | - | - | - | - | - | - | - | - | E | A | T | P | A | D | V | Y | N | T | D | A | T | L | V | L | A | P | - | - | - | G | K | T | N | M | G | R | V | D | A | I | D | L | A | T | G | E | T | K | W | S | Y | E | T | R | A | A | L | Y | D | P | V | L | T | T | G | G | D | L | V | F | V | G | G | I | D | R | D | F | R | A | L | D | A | E | S | G | K | E | V | W | S | T | R | L | P | G | A | V | S | G | Y | T | T | S | Y | S | I | D | G | R | Q | Y | V | A | V | V | S | G | - | G | S | L | - | - | - | - | - | - | - | - | - | - | - | - | - | G | G | P | T | F | G | P | T | T | P | D | V | - | D | S | A | S | G | A | N | G | I | Y | V | F | A | L | P | E | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | A | V | S | N | E | E | I | L | Q | D | P | K | N | P | Q | Q | I | V | T | N | G | L | G | V | Q | G | Q | R | Y | S | P | L | D | L | L | N | V | N | N | V | K | E | L | R | P | V | W | A | F | S | F | G | G | Q | K | Q | R | G | Q | Q | A | Q | P | L | I | K | D | G | V | M | Y | L | T | G | S | Y | - | S | R | V | F | A | V | D | A | R | T | G | - | K | K | L | W | Q | Y | D | A | R | L | P | D | D | I | - | - | R | P | C | C | D | V | I | N | R | G | V | A | L | Y | G | N | L | V | F | F | G | T | L | D | A | K | L | V | A | L | N | K | D | T | G | K | V | V | W | S | K | K | V | A | D | - | H | K | E | G | Y | S | I | S | A | A | P | M | I | V | N | G | K | L | I | T | G | V | A | G | G | E | F | G | V | V | G | K | I | Q | A | Y | N | P | E | N | G | E | L | L | W | M | R | P | T | V | E | G | H | - | - | M | G | Y | V | Y | K | D | G | K | A | I | E | N | G | I | S | G | - | - | - | - | - | G | E | A | G | K | T | W | P | G | - | - | D | L | W | K | - | T | G | G | A | A | P | W | L | G | G | Y | Y | D | P | E | T | N | L | I | L | F | G | T | G | N | P | A | P | W | N | S | H | L | R | P | - | - | - | G | D | N | L | Y | S | S | S | R | L | A | L | N | P | D | D | G | T | I | K | W | H | F | Q | S | T | P | H | D | G | W | D | F | D | G | V | N | E | L | I | S | F | N | Y | K | D | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | K | E | V | K | A | A | A | T | A | D | - | R | N | G | F | F | Y | V | L | D | R | T | N | G | K | F | I | R | G | F | P | F | V | D | K | I | T | W | A | T | G | L | D | - | K | D | G | R | P | I | Y | N | D | A | S | R | P | G | A | P | G | S | E | A | K | G | S | S | V | F | V | A | P | A | F | L | G | A | K | N | W | M | P | M | A | Y | N | K | D | T | G | L | F | Y | V | P | S | N | E | W | G | M | D | I | W | N | E | - | G | I | A | Y | K | - | - | - | - | - | - | - | - | - | K | G | A | A | F | L | G | A | G | F | T | I | K | P | L | N | E | - | - | - | - | D | Y | I | G | V | L | R | A | I | D | P | V | S | G | K | E | V | W | R | H | K | N | Y | A | P | L | W | G | G | V | L | T | T | K | G | N | L | V | F | T | G | T | P | E | G | F | L | Q | A | F | N | A | K | T | G | D | K | V | W | E | F | Q | T | G | S | G | V | L | G | S | P | V | T | W | E | M | D | G | E | Q | Y | V | S | V | V | S | G | W | G | G | A | V | P | - | - | - | - | - | - | - | - | - | - | - | - | - | L | W | G | G | E | V | A | K | R | V | K | D | - | F | N | Q | G | G | M | L | W | T | F | K | L | P | - | - | K | Q | L | Q | Q |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1kb0a2 | A | Q46444 | GO:0005509 |
| d1kb0a2 | A | Q46444 | GO:0009055 |
| d1kb0a2 | A | Q46444 | GO:0016020 |
| d1kb0a2 | A | Q46444 | GO:0016491 |
| d1kb0a2 | A | Q46444 | GO:0016614 |
| d1kb0a2 | A | Q46444 | GO:0020037 |
| d1kb0a2 | A | Q46444 | GO:0030288 |
| d1kb0a2 | A | Q46444 | GO:0042597 |
| d1kb0a2 | A | Q46444 | GO:0046872 |
